ZFLNCG06699
Basic Information
Chromesome: chr11
Start: 12532721
End: 12533015
Transcript: ZFLNCT10261
Known as: ENSDARG00000087368
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 7858.30 |
| SRR800046 | sphere stage | 5azaCyD treatment | 5558.98 |
| SRR800043 | sphere stage | normal | 4850.62 |
| SRR800049 | sphere stage | control treatment | 3910.52 |
| SRR800045 | muscle | normal | 124.10 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 23.87 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 7.86 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 7.02 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 6.23 |
| SRR372791 | dome | normal | 5.68 |
Express in tissues
Correlated coding gene
Download
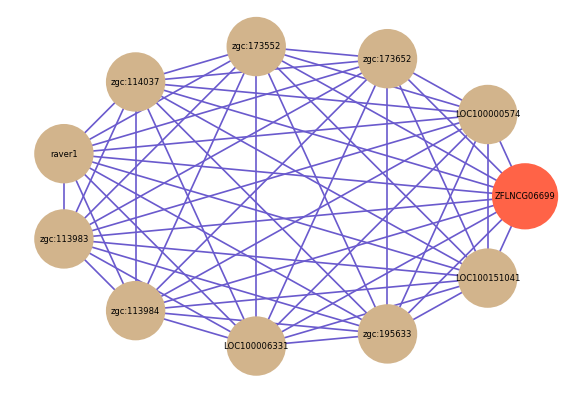
| gene | correlation coefficent |
|---|---|
| LOC100000574 | 0.64 |
| zgc:173652 | 0.64 |
| zgc:173552 | 0.63 |
| zgc:114037 | 0.63 |
| raver1 | 0.62 |
| zgc:113983 | 0.62 |
| zgc:113984 | 0.62 |
| LOC100006331 | 0.62 |
| zgc:195633 | 0.62 |
| LOC100151041 | 0.61 |
Gene Ontology
Download
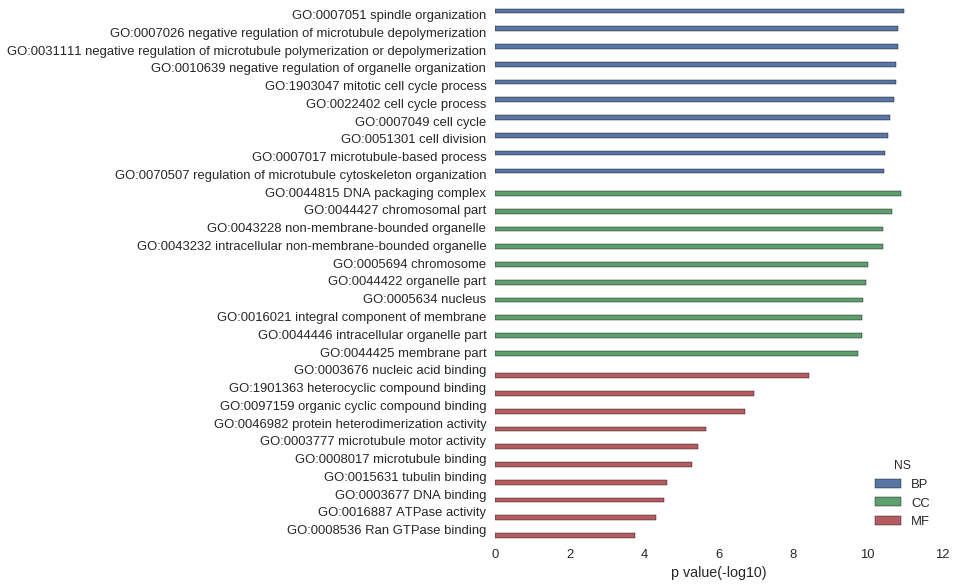
| GO | P value |
|---|---|
| GO:0007051 | 1.02e-11 |
| GO:0007026 | 1.48e-11 |
| GO:0031111 | 1.48e-11 |
| GO:0010639 | 1.69e-11 |
| GO:1903047 | 1.71e-11 |
| GO:0022402 | 1.90e-11 |
| GO:0007049 | 2.41e-11 |
| GO:0051301 | 2.72e-11 |
| GO:0007017 | 3.31e-11 |
| GO:0070507 | 3.59e-11 |
| GO:0044815 | 1.24e-11 |
| GO:0044427 | 2.12e-11 |
| GO:0043228 | 3.84e-11 |
| GO:0043232 | 3.84e-11 |
| GO:0005694 | 9.41e-11 |
| GO:0044422 | 1.08e-10 |
| GO:0005634 | 1.32e-10 |
| GO:0016021 | 1.36e-10 |
| GO:0044446 | 1.38e-10 |
| GO:0044425 | 1.81e-10 |
| GO:0003676 | 3.74e-09 |
| GO:1901363 | 1.14e-07 |
| GO:0097159 | 1.90e-07 |
| GO:0046982 | 2.24e-06 |
| GO:0003777 | 3.54e-06 |
| GO:0008017 | 5.31e-06 |
| GO:0015631 | 2.37e-05 |
| GO:0003677 | 2.95e-05 |
| GO:0016887 | 4.92e-05 |
| GO:0008536 | 1.79e-04 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06699
GTCTGCGTCTCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTTACAGGCGCGATCCCACTACTGATC
AGCACGGGGGATTTGACCTGCTCCGTCTCCGACCTGGGTCGGTTCACCCCTCCTTAAACGACCTGGTGGT
CCCGAGCTCCCCCAGGAGAACCATATCGATACTGAACTTAGCGCGGACACCCGATCAACATAGTCCACTA
TAGCCCAGAAGCCCTGAGCTCAAGCGACCCGCAGGCTCAGCTTCCCAGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC
GTCTGCGTCTCTCTGTGTCGCTCAGGCTGCACTGCAGTGTCTATTTACAGGCGCGATCCCACTACTGATC
AGCACGGGGGATTTGACCTGCTCCGTCTCCGACCTGGGTCGGTTCACCCCTCCTTAAACGACCTGGTGGT
CCCGAGCTCCCCCAGGAGAACCATATCGATACTGAACTTAGCGCGGACACCCGATCAACATAGTCCACTA
TAGCCCAGAAGCCCTGAGCTCAAGCGACCCGCAGGCTCAGCTTCCCAGTAGCTTGGATTACAGGCGCGCG
CCACTGAACCCGGC