ZFLNCG06715
Basic Information
Chromesome: chr11
Start: 12717744
End: 12718038
Transcript: ZFLNCT10278
Known as: ENSDARG00000090238
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800043 | sphere stage | normal | 121.60 |
| SRR800046 | sphere stage | 5azaCyD treatment | 91.83 |
| SRR800037 | egg | normal | 81.18 |
| SRR800049 | sphere stage | control treatment | 36.83 |
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 11.93 |
| SRR748488 | sperm | normal | 11.80 |
| SRR535986 | larvae | nhsl1b mut | 10.45 |
| SRR800045 | muscle | normal | 7.92 |
| SRR527844 | 24hpf | normal | 6.35 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 4.24 |
Express in tissues
Correlated coding gene
Download
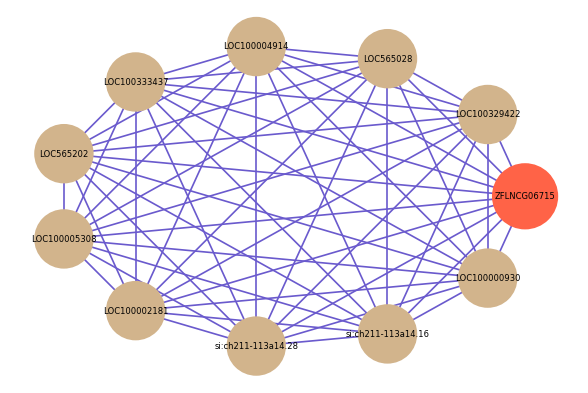
| gene | correlation coefficent |
|---|---|
| LOC100329422 | 0.65 |
| LOC565028 | 0.63 |
| LOC100004914 | 0.63 |
| LOC100333437 | 0.63 |
| LOC565202 | 0.62 |
| LOC100005308 | 0.62 |
| LOC100002181 | 0.60 |
| si:ch211-113a14.28 | 0.60 |
| si:ch211-113a14.16 | 0.60 |
| LOC100000930 | 0.60 |
Gene Ontology
Download
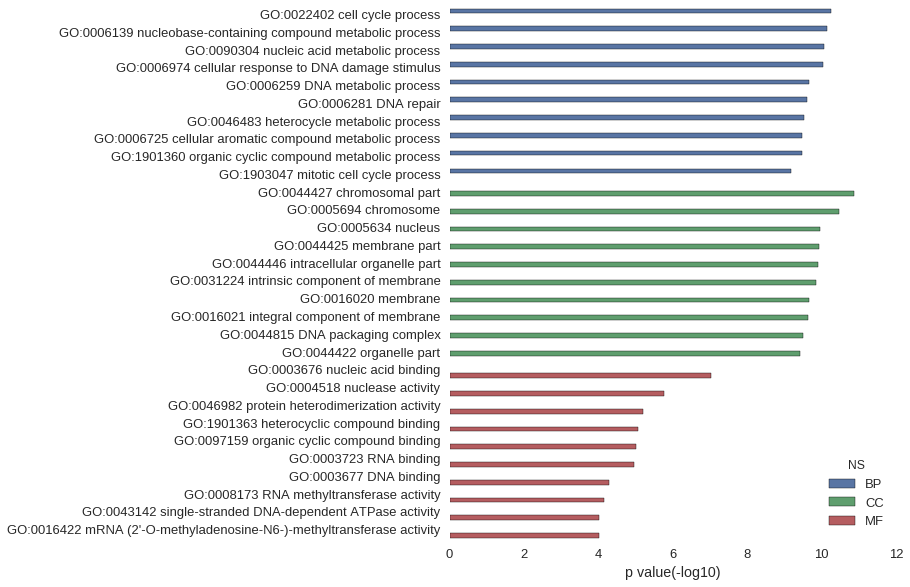
| GO | P value |
|---|---|
| GO:0022402 | 5.70e-11 |
| GO:0006139 | 7.23e-11 |
| GO:0090304 | 8.81e-11 |
| GO:0006974 | 9.36e-11 |
| GO:0006259 | 2.21e-10 |
| GO:0006281 | 2.40e-10 |
| GO:0046483 | 2.92e-10 |
| GO:0006725 | 3.34e-10 |
| GO:1901360 | 3.44e-10 |
| GO:1903047 | 6.44e-10 |
| GO:0044427 | 1.34e-11 |
| GO:0005694 | 3.40e-11 |
| GO:0005634 | 1.09e-10 |
| GO:0044425 | 1.16e-10 |
| GO:0044446 | 1.26e-10 |
| GO:0031224 | 1.39e-10 |
| GO:0016020 | 2.16e-10 |
| GO:0016021 | 2.37e-10 |
| GO:0044815 | 3.12e-10 |
| GO:0044422 | 3.79e-10 |
| GO:0003676 | 9.37e-08 |
| GO:0004518 | 1.70e-06 |
| GO:0046982 | 6.28e-06 |
| GO:1901363 | 8.61e-06 |
| GO:0097159 | 9.53e-06 |
| GO:0003723 | 1.09e-05 |
| GO:0003677 | 5.21e-05 |
| GO:0008173 | 7.21e-05 |
| GO:0043142 | 9.83e-05 |
| GO:0016422 | 9.83e-05 |
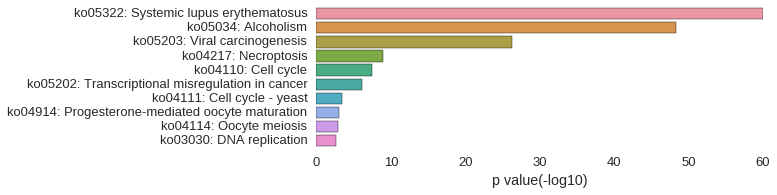
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06715
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGAATTGCTTGAGCTCAG
GGCTTCTGGACTGCAGTGGACTATGTTGATCAGGTGTCCACACTAAGTTCGGTATCGATATGGTGCTCCT
GGGGGAGCTTGGGACCACCAGGTCGTGTAATGAGGGGTGAACCGGCCCAGGTCGGCGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCACACCTGAGAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGAATTGCTTGAGCTCAG
GGCTTCTGGACTGCAGTGGACTATGTTGATCAGGTGTCCACACTAAGTTCGGTATCGATATGGTGCTCCT
GGGGGAGCTTGGGACCACCAGGTCGTGTAATGAGGGGTGAACCGGCCCAGGTCGGCGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCACACCTGAGAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT