ZFLNCG06717
Basic Information
Chromesome: chr11
Start: 12737664
End: 12737958
Transcript: ZFLNCT10280
Known as: ENSDARG00000086624
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800046 | sphere stage | 5azaCyD treatment | 3336.80 |
| SRR800037 | egg | normal | 2605.83 |
| SRR800043 | sphere stage | normal | 2551.27 |
| SRR800049 | sphere stage | control treatment | 2314.62 |
| SRR800045 | muscle | normal | 102.97 |
| SRR748488 | sperm | normal | 22.75 |
| SRR527844 | 24hpf | normal | 6.63 |
| SRR658539 | bud | Gata5/6 morphant | 5.78 |
| SRR801556 | embryo | RPS19 and p53 morpholino | 4.50 |
| SRR527836 | 24hpf | normal | 3.47 |
Express in tissues
Correlated coding gene
Download
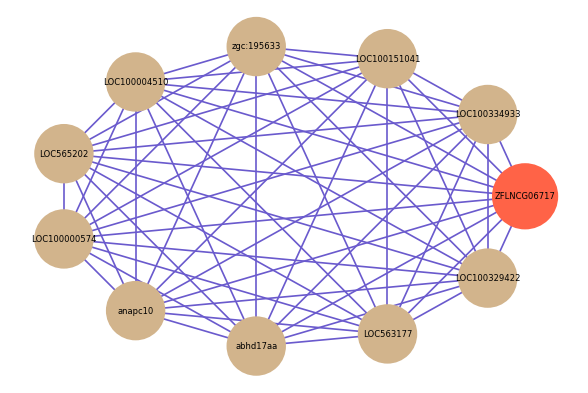
| gene | correlation coefficent |
|---|---|
| LOC100334933 | 0.56 |
| LOC100151041 | 0.56 |
| zgc:195633 | 0.54 |
| LOC100004510 | 0.54 |
| LOC565202 | 0.53 |
| LOC100000574 | 0.53 |
| anapc10 | 0.53 |
| abhd17aa | 0.53 |
| LOC563177 | 0.52 |
| LOC100329422 | 0.52 |
Gene Ontology
Download
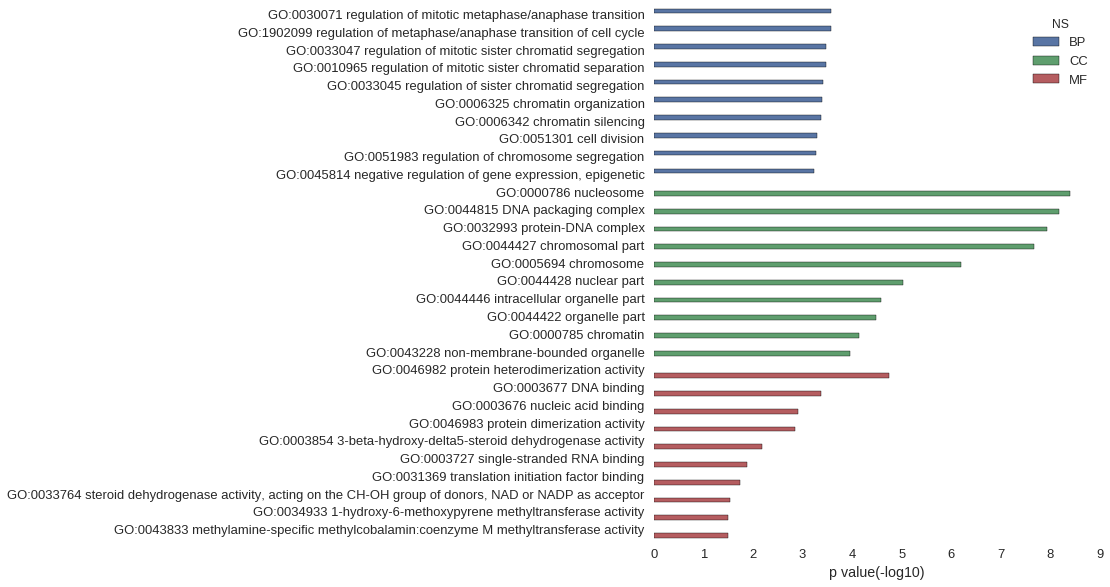
| GO | P value |
|---|---|
| GO:0030071 | 2.65e-04 |
| GO:1902099 | 2.65e-04 |
| GO:0033047 | 3.42e-04 |
| GO:0010965 | 3.42e-04 |
| GO:0033045 | 3.85e-04 |
| GO:0006325 | 4.01e-04 |
| GO:0006342 | 4.30e-04 |
| GO:0051301 | 5.09e-04 |
| GO:0051983 | 5.27e-04 |
| GO:0045814 | 5.79e-04 |
| GO:0000786 | 4.07e-09 |
| GO:0044815 | 6.77e-09 |
| GO:0032993 | 1.20e-08 |
| GO:0044427 | 2.13e-08 |
| GO:0005694 | 6.35e-07 |
| GO:0044428 | 9.26e-06 |
| GO:0044446 | 2.66e-05 |
| GO:0044422 | 3.23e-05 |
| GO:0000785 | 7.11e-05 |
| GO:0043228 | 1.08e-04 |
| GO:0046982 | 1.84e-05 |
| GO:0003677 | 4.30e-04 |
| GO:0003676 | 1.25e-03 |
| GO:0046983 | 1.43e-03 |
| GO:0003854 | 6.52e-03 |
| GO:0003727 | 1.30e-02 |
| GO:0031369 | 1.78e-02 |
| GO:0033764 | 2.90e-02 |
| GO:0034933 | 3.22e-02 |
| GO:0043833 | 3.22e-02 |
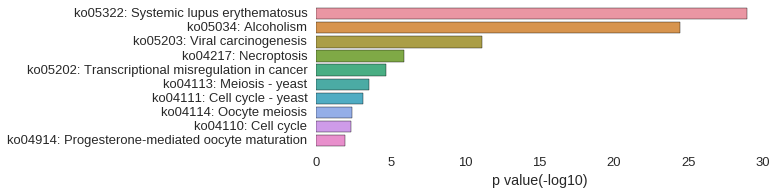
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06717
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGGCTGCAGTGGACTATGTTGATCGGGTGTCCGCGTTAAGTTCGGTATCGGTATGGTTCTCCT
GGGGGAGCTCGGGACCACCAGGTCGTTTACGGAGGGGTGAACCGGCCCAGGTTAGAGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCGCGCCTGTAAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT
CCGGGTTCAGTGGCGCGCGCCTGTAATCCAAGCTACTGGGAGGCTGAGGCTGCGGATCGCTTGAGCTCAG
GGCTTCTGGGCTGCAGTGGACTATGTTGATCGGGTGTCCGCGTTAAGTTCGGTATCGGTATGGTTCTCCT
GGGGGAGCTCGGGACCACCAGGTCGTTTACGGAGGGGTGAACCGGCCCAGGTTAGAGACGGAGCAGGTCA
AATCCCCCGTGCTGATCAGTAGTGGGATCGCGCCTGTAAATAGACACTGCAGTGCAGCCTGAGCGACACA
GAGAGACGCAGACT