ZFLNCG06729
Basic Information
Chromesome: chr11
Start: 14322585
End: 14323222
Transcript: ZFLNCT10292
Known as: ENSDARG00000097955
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516132 | skin | male and 5 month | 3.41 |
| SRR1562529 | intestine and pancreas | normal | 1.87 |
| SRR516126 | skin | male and 5 month | 1.56 |
| SRR516129 | skin | male and 5 month | 1.53 |
| SRR516131 | skin | male and 5 month | 1.43 |
| SRR941749 | anterior pectoral fin | normal | 1.30 |
| SRR1205154 | 5dpf | transgenic sqET20 | 1.06 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 1.03 |
| SRR516135 | skin | male and 5 month | 1.03 |
| SRR594769 | blood | normal | 0.99 |
Express in tissues
Correlated coding gene
Download
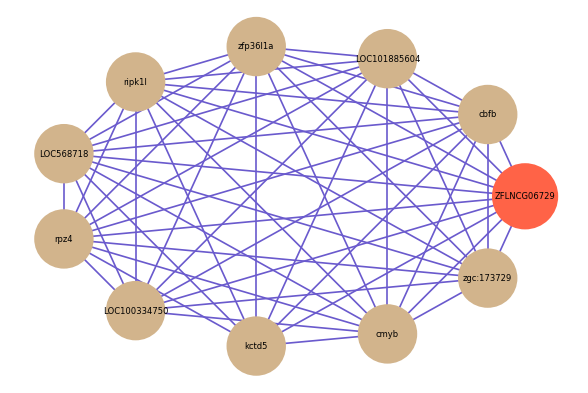
| gene | correlation coefficent |
|---|---|
| cbfb | 0.56 |
| LOC101885604 | 0.54 |
| zfp36l1a | 0.54 |
| ripk1l | 0.54 |
| LOC568718 | 0.54 |
| rpz4 | 0.53 |
| LOC100334750 | 0.53 |
| kctd5 | 0.53 |
| cmyb | 0.52 |
| zgc:173729 | 0.51 |
Gene Ontology
Download
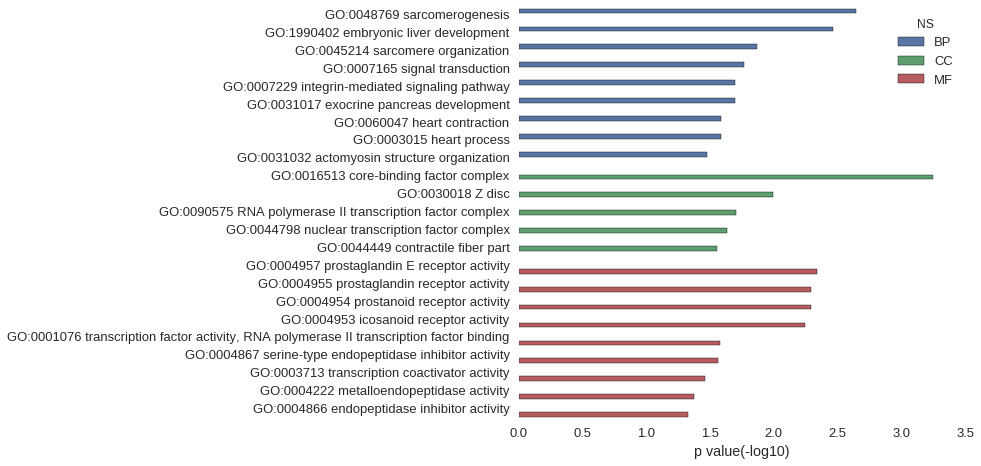
| GO | P value |
|---|---|
| GO:0048769 | 2.27e-03 |
| GO:1990402 | 3.41e-03 |
| GO:0045214 | 1.36e-02 |
| GO:0007165 | 1.72e-02 |
| GO:0007229 | 2.03e-02 |
| GO:0031017 | 2.03e-02 |
| GO:0060047 | 2.59e-02 |
| GO:0003015 | 2.59e-02 |
| GO:0031032 | 3.31e-02 |
| GO:0016513 | 5.69e-04 |
| GO:0030018 | 1.02e-02 |
| GO:0090575 | 1.97e-02 |
| GO:0044798 | 2.31e-02 |
| GO:0044449 | 2.81e-02 |
| GO:0004957 | 4.54e-03 |
| GO:0004955 | 5.11e-03 |
| GO:0004954 | 5.11e-03 |
| GO:0004953 | 5.67e-03 |
| GO:0001076 | 2.64e-02 |
| GO:0004867 | 2.75e-02 |
| GO:0003713 | 3.47e-02 |
| GO:0004222 | 4.24e-02 |
| GO:0004866 | 4.73e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06729
GCCAGAGGTCACCAAGCTTGAACTTTGCAACACAGCAAACTGCGAAACTTAGCAGGAATATGACTGGAAG
TCTAAGGGGAGAAAAGTACAGTGTGAACGCAGCTTGAGTGTGTTAAAGCAGTGTTCAAACTAAACTGAGC
AGGGCTGCAGCCCTCCAGGAAGAGTTTGACACCCCTAAGGTAACTAAACGCTTTATTATACTAAATGCCT
TATTCTAACTGATGTGATGACATCACACGCATAATCTTAACATCAATAATAACTACATTGTAGTAAAAGT
ACTCACAAAGATAACCATTATTACAAAAGCTGCCAGGTAGGAGGTACGAGCCATCCACATGCTGACAAAA
CGATAGTGTTCTCCAGACACCACGTTCCTGAGGAAGCCTAAAAGCACACAATCAATCTCTGACATTGTAA
TTATGCACTCAAGAAGGAAGCACTGATCTTTTGGGAGAACACGTAATTGTTTACCTTTGTTTTCCTCATT
CTCTGCTAATGCCTTCACACTGGACATTAGAATATCATCATAACCCAGGCACTCGTCCAGGAGGAAGCGA
CTGAAACCATCTCCAAAGCACTGATCTTTCATTGGATCTACAGACAAGCAGAGAGAGATTGACAGAGATT
CACAATG
GCCAGAGGTCACCAAGCTTGAACTTTGCAACACAGCAAACTGCGAAACTTAGCAGGAATATGACTGGAAG
TCTAAGGGGAGAAAAGTACAGTGTGAACGCAGCTTGAGTGTGTTAAAGCAGTGTTCAAACTAAACTGAGC
AGGGCTGCAGCCCTCCAGGAAGAGTTTGACACCCCTAAGGTAACTAAACGCTTTATTATACTAAATGCCT
TATTCTAACTGATGTGATGACATCACACGCATAATCTTAACATCAATAATAACTACATTGTAGTAAAAGT
ACTCACAAAGATAACCATTATTACAAAAGCTGCCAGGTAGGAGGTACGAGCCATCCACATGCTGACAAAA
CGATAGTGTTCTCCAGACACCACGTTCCTGAGGAAGCCTAAAAGCACACAATCAATCTCTGACATTGTAA
TTATGCACTCAAGAAGGAAGCACTGATCTTTTGGGAGAACACGTAATTGTTTACCTTTGTTTTCCTCATT
CTCTGCTAATGCCTTCACACTGGACATTAGAATATCATCATAACCCAGGCACTCGTCCAGGAGGAAGCGA
CTGAAACCATCTCCAAAGCACTGATCTTTCATTGGATCTACAGACAAGCAGAGAGAGATTGACAGAGATT
CACAATG