ZFLNCG06920
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 335.17 |
| SRR592701 | pineal gland | normal | 142.72 |
| SRR1028002 | head | normal | 133.50 |
| SRR592703 | pineal gland | normal | 113.59 |
| SRR592702 | pineal gland | normal | 106.57 |
| SRR1028004 | head | normal | 98.91 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 96.62 |
| SRR516122 | skin | male and 3.5 year | 78.67 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 78.39 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 72.67 |
Express in tissues
Correlated coding gene
Download
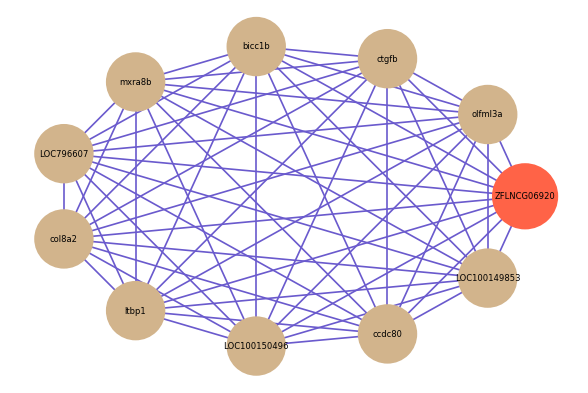
| gene | correlation coefficent |
|---|---|
| olfml3a | 0.78 |
| ctgfb | 0.77 |
| bicc1b | 0.76 |
| mxra8b | 0.75 |
| LOC796607 | 0.75 |
| col8a2 | 0.74 |
| ltbp1 | 0.74 |
| LOC100150496 | 0.73 |
| ccdc80 | 0.73 |
| LOC100149853 | 0.72 |
Gene Ontology
Download
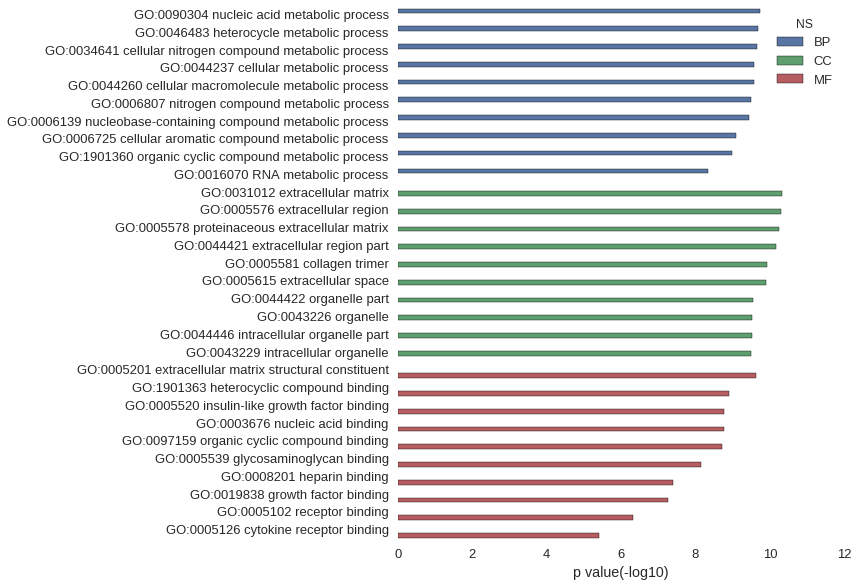
| GO | P value |
|---|---|
| GO:0090304 | 1.89e-10 |
| GO:0046483 | 2.02e-10 |
| GO:0034641 | 2.26e-10 |
| GO:0044237 | 2.67e-10 |
| GO:0044260 | 2.73e-10 |
| GO:0006807 | 3.16e-10 |
| GO:0006139 | 3.52e-10 |
| GO:0006725 | 8.32e-10 |
| GO:1901360 | 1.06e-09 |
| GO:0016070 | 4.62e-09 |
| GO:0031012 | 4.79e-11 |
| GO:0005576 | 4.90e-11 |
| GO:0005578 | 5.81e-11 |
| GO:0044421 | 6.65e-11 |
| GO:0005581 | 1.18e-10 |
| GO:0005615 | 1.23e-10 |
| GO:0044422 | 2.83e-10 |
| GO:0043226 | 3.04e-10 |
| GO:0044446 | 3.09e-10 |
| GO:0043229 | 3.29e-10 |
| GO:0005201 | 2.37e-10 |
| GO:1901363 | 1.26e-09 |
| GO:0005520 | 1.66e-09 |
| GO:0003676 | 1.66e-09 |
| GO:0097159 | 1.94e-09 |
| GO:0005539 | 6.99e-09 |
| GO:0008201 | 4.02e-08 |
| GO:0019838 | 5.59e-08 |
| GO:0005102 | 4.90e-07 |
| GO:0005126 | 3.82e-06 |
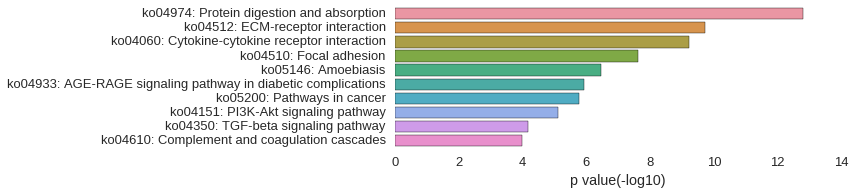
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06920
CTGAAATCATGTGCTTTGAGTTGCTTTTGCACTGTTTAGTTTAGCGCTGCTTTAAGCTCATAGCATATTT
ACACTTTTAGATGTACACGAGGGTCTGACACACAAAAATGAACTGCTTTTCAATCCTCTGGTGTCAACAC
TGAACCGGCCTGTTATTTCAGTTTTTGGAAAGAAACTAAGACTGAACAAAAGCAACACAAGACAACACTG
AAAATGGCAAAAATAGCCATTGACAAGTG
CTGAAATCATGTGCTTTGAGTTGCTTTTGCACTGTTTAGTTTAGCGCTGCTTTAAGCTCATAGCATATTT
ACACTTTTAGATGTACACGAGGGTCTGACACACAAAAATGAACTGCTTTTCAATCCTCTGGTGTCAACAC
TGAACCGGCCTGTTATTTCAGTTTTTGGAAAGAAACTAAGACTGAACAAAAGCAACACAAGACAACACTG
AAAATGGCAAAAATAGCCATTGACAAGTG