ZFLNCG06925
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562529 | intestine and pancreas | normal | 13.29 |
| SRR516133 | skin | male and 5 month | 10.26 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 8.26 |
| SRR516135 | skin | male and 5 month | 6.28 |
| SRR527834 | head | normal | 5.31 |
| ERR023146 | head kidney | normal | 5.03 |
| SRR527835 | tail | normal | 4.81 |
| SRR941749 | anterior pectoral fin | normal | 4.21 |
| ERR023145 | heart | normal | 4.11 |
| SRR519752 | 7 dpf | VD3 treatment | 4.07 |
Express in tissues
Correlated coding gene
Download
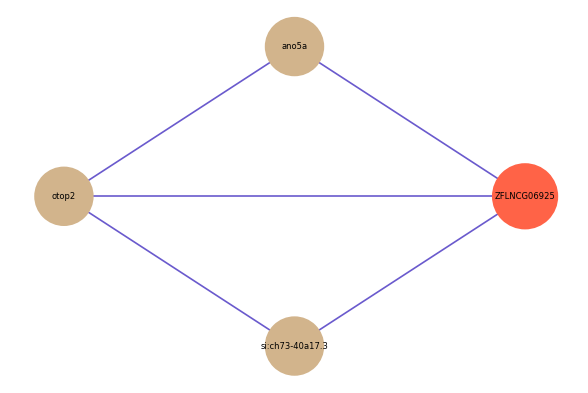
| gene | correlation coefficent |
|---|---|
| ano5a | 0.55 |
| otop2 | 0.54 |
| si:ch73-40a17.3 | 0.50 |
Gene Ontology
Download
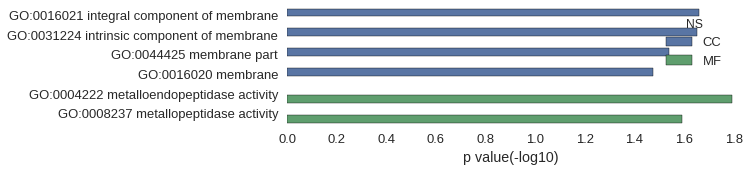
| GO | P value |
|---|---|
| GO:0016021 | 2.19e-02 |
| GO:0031224 | 2.23e-02 |
| GO:0044425 | 2.90e-02 |
| GO:0016020 | 3.34e-02 |
| GO:0004222 | 1.61e-02 |
| GO:0008237 | 2.56e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06925
CAGTAATAGATGATGTATTCTGCTCTTCTGTTGTAGAAGTTAACGGAGATGGAGAGGTACCTGTTGTGTA
TACTGCAGTTTCCGATGTTGAACCACTTTCAGTTGTAGAAGTTTTGATTGTGGTTGCGATCACAGATGAA
TCTTCTGCTGTAAATGTTTGTGTCTGTTGGCTTGTGGTTTGTTCAACAGTAGTTGAAGATATTTCTGTTG
ACAATGTTGATGTGTTCTGTGGTGTTGTGGAATAGGCTGAAAATGAAGTGCTGCCTGTTATACATGATGT
ATTCTGCTCTTTTGTTGTTGAAGTTACCGGAGATGGAAAGGTACCTGTTGTGTATAATGTAGTTTCCGAT
GTAGAAGATTTAAATGTGGTTGTAGTCCTAGGGGACTCTTCTGTTGTGAAGGGTGTAGATTCTGTTATTG
TGGGTGAACTATCAGGACTGGTTTTTGTCTGTTGGCGTGTGGTAAGTTCTACAGTAGCTGAAGTTTCTGT
TGTAAACTCAGTCATAGATGATGTAGTCTGCTCTTCTGTTGTAGAAGTTACCGGAGATGGAGAGGTACCT
GTTGTGTATAGTGTAGTTTCCATCGTA
CAGTAATAGATGATGTATTCTGCTCTTCTGTTGTAGAAGTTAACGGAGATGGAGAGGTACCTGTTGTGTA
TACTGCAGTTTCCGATGTTGAACCACTTTCAGTTGTAGAAGTTTTGATTGTGGTTGCGATCACAGATGAA
TCTTCTGCTGTAAATGTTTGTGTCTGTTGGCTTGTGGTTTGTTCAACAGTAGTTGAAGATATTTCTGTTG
ACAATGTTGATGTGTTCTGTGGTGTTGTGGAATAGGCTGAAAATGAAGTGCTGCCTGTTATACATGATGT
ATTCTGCTCTTTTGTTGTTGAAGTTACCGGAGATGGAAAGGTACCTGTTGTGTATAATGTAGTTTCCGAT
GTAGAAGATTTAAATGTGGTTGTAGTCCTAGGGGACTCTTCTGTTGTGAAGGGTGTAGATTCTGTTATTG
TGGGTGAACTATCAGGACTGGTTTTTGTCTGTTGGCGTGTGGTAAGTTCTACAGTAGCTGAAGTTTCTGT
TGTAAACTCAGTCATAGATGATGTAGTCTGCTCTTCTGTTGTAGAAGTTACCGGAGATGGAGAGGTACCT
GTTGTGTATAGTGTAGTTTCCATCGTA

