ZFLNCG06943
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 52.93 |
| SRR1104059 | heart | kctd10 mut | 34.90 |
| SRR1562531 | muscle | normal | 30.87 |
| ERR145648 | skeletal muscle | 27 degree_C to 27 degree_C | 26.33 |
| SRR891510 | muscle | normal | 26.32 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 19.84 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 19.10 |
| SRR1104058 | heart | normal | 16.96 |
| SRR514030 | retina | id2a morpholino | 16.87 |
| SRR594771 | endothelium | normal | 15.73 |
Express in tissues
Correlated coding gene
Download
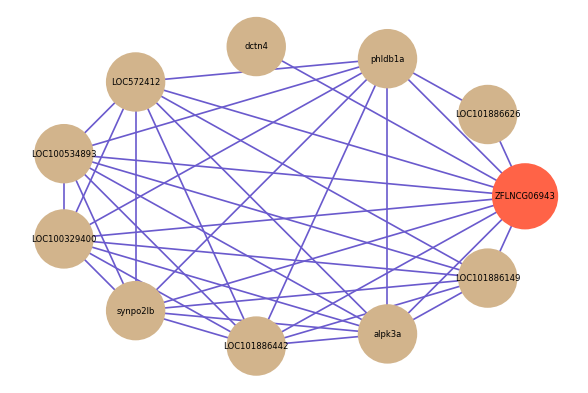
| gene | correlation coefficent |
|---|---|
| LOC101886626 | 0.69 |
| dctn4 | 0.57 |
| LOC572412 | 0.56 |
| LOC100534893 | 0.55 |
| LOC100329400 | 0.54 |
| phldb1a | 0.51 |
| synpo2lb | 0.51 |
| LOC101886442 | 0.51 |
| alpk3a | 0.51 |
| LOC101886149 | 0.51 |
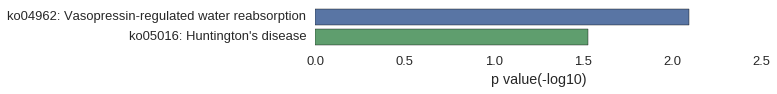
Gene Ontology
Download
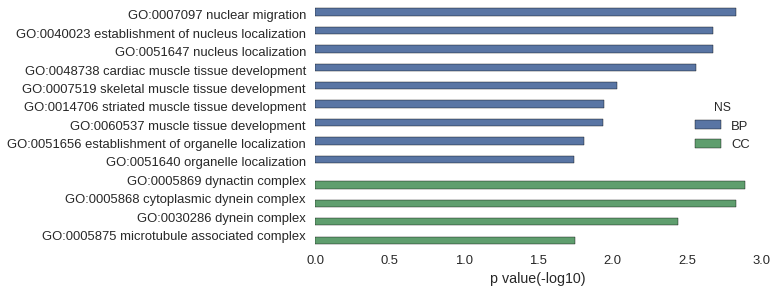
| GO | P value |
|---|---|
| GO:0007097 | 1.49e-03 |
| GO:0040023 | 2.13e-03 |
| GO:0051647 | 2.13e-03 |
| GO:0048738 | 2.77e-03 |
| GO:0007519 | 9.35e-03 |
| GO:0014706 | 1.15e-02 |
| GO:0060537 | 1.17e-02 |
| GO:0051656 | 1.57e-02 |
| GO:0051640 | 1.82e-02 |
| GO:0005869 | 1.28e-03 |
| GO:0005868 | 1.49e-03 |
| GO:0030286 | 3.62e-03 |
| GO:0005875 | 1.80e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG06943
TTGTCTGTTTGTGTCTCTTTTCCAGGAAGTATTGGCTCAGTAAAGTGGGCAGCCCGAGCTCTCGCATCGC
CCGCGCTGAGTTCAGCAACGAGAGAGACATTTCCAAACTGGAGATTCATGGATTTGGCACGCGCTACTAA
ACAAACACACAAAACCACAACACGACAAACTCACAACACAACACAACAAACCACAACACAACAAACTTAA
AACACACAAAACCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACAAC
ACAACAAACCACAACACAACAAACTTAAAACACACAAAACTACAACACAACAAACTCTCAACACACAAAA
CCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACAACA
CGACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACAACACAACAAAC
TCACAACACACAAAACCACAACATAACAAACTCACAACACACAAAACCACAACACAACAAACTCACAACA
CACAAAAACCAGAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCTCAACACACAAAA
ACCAGAACACAACAAACTCACAACACAACACAACAAACTATAACACAACAAACTCACAACACAACACACA
AAACCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACA
ACACAACACATATTTGGTAGAGATTAACACACTTTACATAGAAAACAACTCCTTTGTCCCTTTATTGTCC
AGTTTTGCTTCTATTTGAATTGTATTACTGTAAATATGAACCTAACCTTGATTATTTGTAAAGTTTACTT
AAAGTATAAGTGATTTAATATTTTAATTAATTAAGCATTTTTAAAAGGATGCATTTATTTGAACAAATAT
AGATGAAATGTTTTTTTTTTACAATTTAATTAATTTCTTAACATTATTATATTTTAAAAGGTATTTATTC
CTGTAATAGCTGAAACAATTGTTTCATAAATATAAATAAAACATAAATACTCAC
TTGTCTGTTTGTGTCTCTTTTCCAGGAAGTATTGGCTCAGTAAAGTGGGCAGCCCGAGCTCTCGCATCGC
CCGCGCTGAGTTCAGCAACGAGAGAGACATTTCCAAACTGGAGATTCATGGATTTGGCACGCGCTACTAA
ACAAACACACAAAACCACAACACGACAAACTCACAACACAACACAACAAACCACAACACAACAAACTTAA
AACACACAAAACCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACAAC
ACAACAAACCACAACACAACAAACTTAAAACACACAAAACTACAACACAACAAACTCTCAACACACAAAA
CCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACAACA
CGACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACAACACAACAAAC
TCACAACACACAAAACCACAACATAACAAACTCACAACACACAAAACCACAACACAACAAACTCACAACA
CACAAAAACCAGAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCTCAACACACAAAA
ACCAGAACACAACAAACTCACAACACAACACAACAAACTATAACACAACAAACTCACAACACAACACACA
AAACCACAACACAACAAACTCACAACACACAAAACCACAACACGACAAACTCACAACACACAAAACCACA
ACACAACACATATTTGGTAGAGATTAACACACTTTACATAGAAAACAACTCCTTTGTCCCTTTATTGTCC
AGTTTTGCTTCTATTTGAATTGTATTACTGTAAATATGAACCTAACCTTGATTATTTGTAAAGTTTACTT
AAAGTATAAGTGATTTAATATTTTAATTAATTAAGCATTTTTAAAAGGATGCATTTATTTGAACAAATAT
AGATGAAATGTTTTTTTTTTACAATTTAATTAATTTCTTAACATTATTATATTTTAAAAGGTATTTATTC
CTGTAATAGCTGAAACAATTGTTTCATAAATATAAATAAAACATAAATACTCAC