ZFLNCG07005
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516134 | skin | male and 5 month | 144.89 |
| SRR372793 | shield | normal | 109.67 |
| SRR1647684 | spleen | SVCV treatment | 104.83 |
| SRR1299127 | caudal fin | Two days time after treatment | 102.97 |
| ERR023145 | heart | normal | 102.83 |
| SRR1647681 | head kidney | SVCV treatment | 101.26 |
| SRR1299125 | caudal fin | Half day time after treatment | 97.93 |
| SRR1299126 | caudal fin | One day time after treatment | 90.56 |
| SRR1299124 | caudal fin | Zero day time after treatment | 89.01 |
| ERR023143 | swim bladder | normal | 87.57 |
Express in tissues
Correlated coding gene
Download
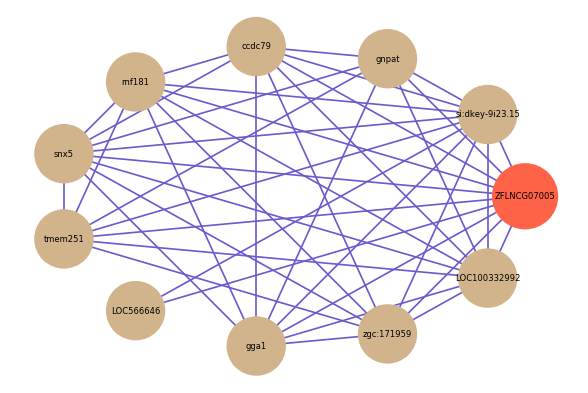
| gene | correlation coefficent |
|---|---|
| si:dkey-9i23.15 | 0.59 |
| gnpat | 0.58 |
| ccdc79 | 0.58 |
| rnf181 | 0.58 |
| snx5 | 0.58 |
| tmem251 | 0.57 |
| LOC566646 | 0.57 |
| gga1 | 0.57 |
| zgc:171959 | 0.57 |
| LOC100332992 | 0.56 |
Gene Ontology
Download
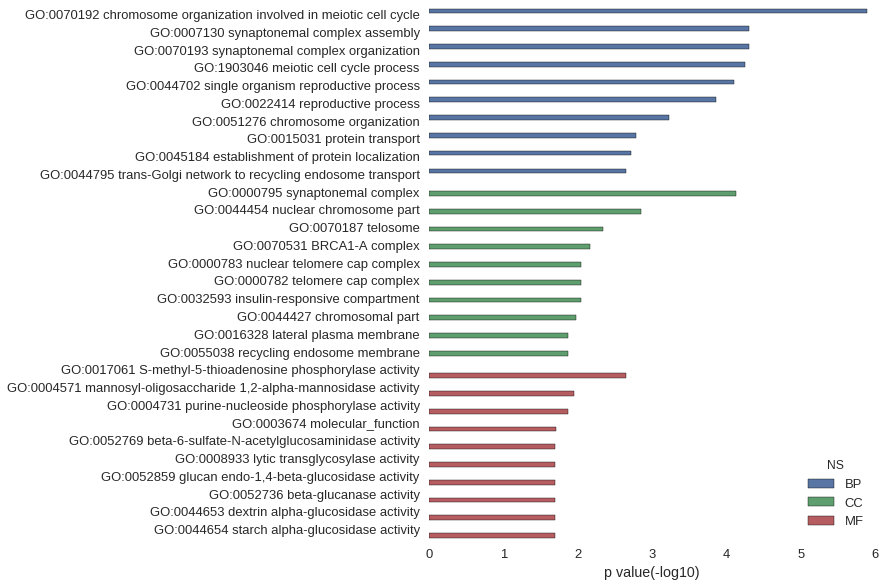
| GO | P value |
|---|---|
| GO:0070192 | 1.27e-06 |
| GO:0007130 | 4.99e-05 |
| GO:0070193 | 4.99e-05 |
| GO:1903046 | 5.56e-05 |
| GO:0044702 | 8.00e-05 |
| GO:0022414 | 1.36e-04 |
| GO:0051276 | 5.96e-04 |
| GO:0015031 | 1.62e-03 |
| GO:0045184 | 1.93e-03 |
| GO:0044795 | 2.27e-03 |
| GO:0000795 | 7.47e-05 |
| GO:0044454 | 1.40e-03 |
| GO:0070187 | 4.54e-03 |
| GO:0070531 | 6.81e-03 |
| GO:0000783 | 9.07e-03 |
| GO:0000782 | 9.07e-03 |
| GO:0032593 | 9.07e-03 |
| GO:0044427 | 1.04e-02 |
| GO:0016328 | 1.36e-02 |
| GO:0055038 | 1.36e-02 |
| GO:0017061 | 2.27e-03 |
| GO:0004571 | 1.13e-02 |
| GO:0004731 | 1.36e-02 |
| GO:0003674 | 1.97e-02 |
| GO:0052769 | 2.03e-02 |
| GO:0008933 | 2.03e-02 |
| GO:0052859 | 2.03e-02 |
| GO:0052736 | 2.03e-02 |
| GO:0044653 | 2.03e-02 |
| GO:0044654 | 2.03e-02 |
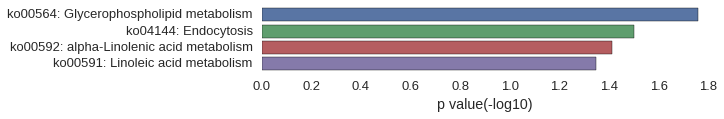
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07005
TGCATTTATTTACTGACTTGAGCTCTATAAAATCAATAGAACAGTGAACAAACTGGCTGTGAATCAGTAG
AAACAGTGATAAAGGCAGAACTGAACAGTGTCACAGTGGTCTGACATCATTAATAAAACAACAACTGCAC
TATTTCAGATTAAATATTCTCATTTAACAGCTCATCAAGACTTAAGTAATTGATTACAGGTGAGTTTTAC
CATTTTTCAATCCACTCAGCCAATCTCAACGTCTGACGGAGCACTTTTAGCTTAGCTTAGCATAGATAAT
GGAATCGGATTAGACCATTAG
TGCATTTATTTACTGACTTGAGCTCTATAAAATCAATAGAACAGTGAACAAACTGGCTGTGAATCAGTAG
AAACAGTGATAAAGGCAGAACTGAACAGTGTCACAGTGGTCTGACATCATTAATAAAACAACAACTGCAC
TATTTCAGATTAAATATTCTCATTTAACAGCTCATCAAGACTTAAGTAATTGATTACAGGTGAGTTTTAC
CATTTTTCAATCCACTCAGCCAATCTCAACGTCTGACGGAGCACTTTTAGCTTAGCTTAGCATAGATAAT
GGAATCGGATTAGACCATTAG