ZFLNCG07062
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 69.15 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 62.60 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 58.01 |
| SRR800043 | sphere stage | normal | 25.68 |
| SRR800037 | egg | normal | 21.37 |
| SRR800049 | sphere stage | control treatment | 19.54 |
| SRR800046 | sphere stage | 5azaCyD treatment | 11.34 |
| SRR1562533 | testis | normal | 4.30 |
| SRR516135 | skin | male and 5 month | 3.95 |
| SRR748488 | sperm | normal | 3.00 |
Express in tissues
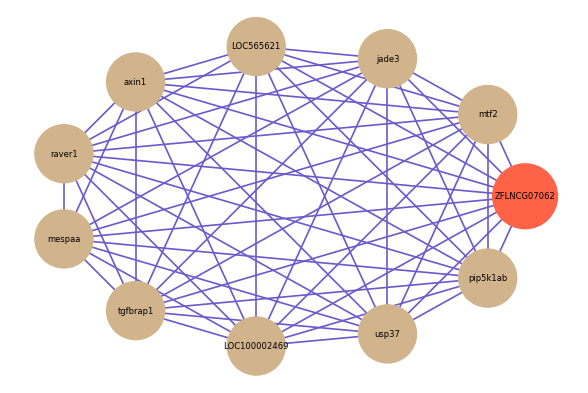
Correlated coding gene
Download
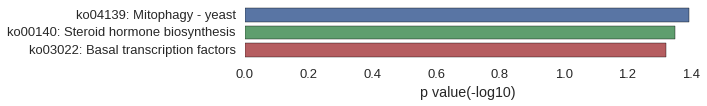
Gene Ontology
Download
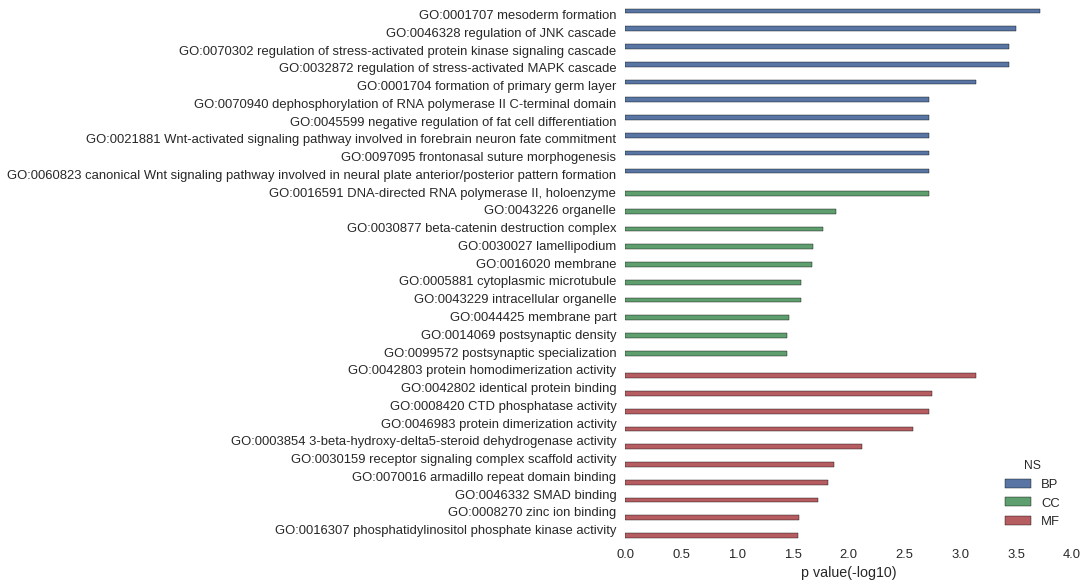
| GO | P value |
|---|---|
| GO:0001707 | 1.93e-04 |
| GO:0046328 | 3.18e-04 |
| GO:0070302 | 3.67e-04 |
| GO:0032872 | 3.67e-04 |
| GO:0001704 | 7.28e-04 |
| GO:0070940 | 1.92e-03 |
| GO:0045599 | 1.92e-03 |
| GO:0021881 | 1.92e-03 |
| GO:0097095 | 1.92e-03 |
| GO:0060823 | 1.92e-03 |
| GO:0016591 | 1.92e-03 |
| GO:0043226 | 1.30e-02 |
| GO:0030877 | 1.71e-02 |
| GO:0030027 | 2.09e-02 |
| GO:0016020 | 2.14e-02 |
| GO:0005881 | 2.65e-02 |
| GO:0043229 | 2.66e-02 |
| GO:0044425 | 3.43e-02 |
| GO:0014069 | 3.59e-02 |
| GO:0099572 | 3.59e-02 |
| GO:0042803 | 7.29e-04 |
| GO:0042802 | 1.79e-03 |
| GO:0008420 | 1.92e-03 |
| GO:0046983 | 2.65e-03 |
| GO:0003854 | 7.65e-03 |
| GO:0030159 | 1.34e-02 |
| GO:0070016 | 1.53e-02 |
| GO:0046332 | 1.90e-02 |
| GO:0008270 | 2.79e-02 |
| GO:0016307 | 2.84e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07062
TCCGACGCCGGTTGATTGTGCTATGACAAGTTTTTACCGTGCAGGGCACTGAAATGATGTGGAACATAAA
AATTAGGATGTTATTGATAGCACTGTTGATAAATTGGGAATTTGTAAAATCGACAATAGCCCATCTTAAC
AAAAACGATATAACTTTAGACACTGACCGAGCCAGTGAGTCTCTCAAAGCCAAGCAGCCTCCGGTAGATG
TGGAAAGATTGCGATTGAGAGTCGACGATGCATATGAGGAATGTAAACGATATCGCAACGCTTCAGCGTC
TTCAGGTGACCCGAACAAAGACTCAATGCCAAAATCCACGTTCCACTGCCATGCATGTTGGCCGGAACAC
AGTTTGGATGAAAAATTAAGGAACGAAGACTCAAGCCAAAGTGGATTGAATCCCCCGCAAAACGACGAAT
CTGAGGATCTTCTCAATGAGCCTGTGACCTGGAGGTATTTGAAAA
TCCGACGCCGGTTGATTGTGCTATGACAAGTTTTTACCGTGCAGGGCACTGAAATGATGTGGAACATAAA
AATTAGGATGTTATTGATAGCACTGTTGATAAATTGGGAATTTGTAAAATCGACAATAGCCCATCTTAAC
AAAAACGATATAACTTTAGACACTGACCGAGCCAGTGAGTCTCTCAAAGCCAAGCAGCCTCCGGTAGATG
TGGAAAGATTGCGATTGAGAGTCGACGATGCATATGAGGAATGTAAACGATATCGCAACGCTTCAGCGTC
TTCAGGTGACCCGAACAAAGACTCAATGCCAAAATCCACGTTCCACTGCCATGCATGTTGGCCGGAACAC
AGTTTGGATGAAAAATTAAGGAACGAAGACTCAAGCCAAAGTGGATTGAATCCCCCGCAAAACGACGAAT
CTGAGGATCTTCTCAATGAGCCTGTGACCTGGAGGTATTTGAAAA