ZFLNCG07130
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 3.65 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 2.29 |
| SRR1562529 | intestine and pancreas | normal | 1.94 |
| SRR1647683 | spleen | SVCV treatment | 0.95 |
| SRR1647682 | spleen | normal | 0.77 |
| SRR1035237 | liver | transgenic UHRF1 | 0.62 |
| SRR1647684 | spleen | SVCV treatment | 0.59 |
| SRR1534349 | larvae | DMSO vehicle | 0.54 |
| SRR726540 | 5 dpf | normal | 0.49 |
| SRR1647679 | head kidney | normal | 0.46 |
Express in tissues
Gene Ontology
Download
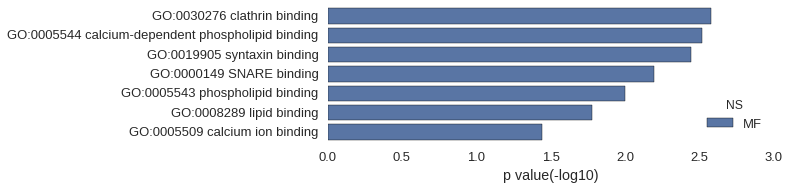
| GO | P value |
|---|---|
| GO:0030276 | 2.63e-03 |
| GO:0005544 | 3.06e-03 |
| GO:0019905 | 3.62e-03 |
| GO:0000149 | 6.40e-03 |
| GO:0005543 | 1.00e-02 |
| GO:0008289 | 1.66e-02 |
| GO:0005509 | 3.62e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07130
CCAGGATATGTTTGACTACATGTTCTTCTATCTTTGTGCACCTCCTTACCAATGGGTATAACACTATTGT
AAACTAGACTGTATGCCTGGGCTACCTTGCTGAATGCCTCAACACAAGTGAAGTATATGCTTGAGGCTTG
GCTACATTTCCAACCAGCATAACGCCAGTGAACTAGACAATGTAAGTCAAGGCTACCTTGTCAACCATTT
ATTCTTTTCTTTTCGGCTCAGTCCCTTTATTAATCAGGGGTTGAATGAACTTGTCAACTTATCATCATGT
GTTTTATGCAGCGGATGCCCTTCCAGGTGCAACCCATTACTGGGAAACACCCCCTTGCCAACCAGCTTAG
CATTAAAATCCACACAATGTGTCCCTGTAAAAAAATCACATAAAAGGTTGAGAGAAATAACAAACCCAGC
AACCATCTGAGAAAGTACAAATGTAAAAAATGGTTATTTCATTGCTATTTGCATAAGTTTCCAAAAACTT
ACCAGGTCTGGAAAATGCCATGTCAAAGTCTTTCCAGGTTTTTCACAACCATAAGAACCCTGTCTGTAGT
CGTGTCATTCTACATCTGTGAGACTTGTTGCATAGTTGTGTGATTCTACATCTGTGAGACTTGTTGCAGT
GCTTGGCGAA
CCAGGATATGTTTGACTACATGTTCTTCTATCTTTGTGCACCTCCTTACCAATGGGTATAACACTATTGT
AAACTAGACTGTATGCCTGGGCTACCTTGCTGAATGCCTCAACACAAGTGAAGTATATGCTTGAGGCTTG
GCTACATTTCCAACCAGCATAACGCCAGTGAACTAGACAATGTAAGTCAAGGCTACCTTGTCAACCATTT
ATTCTTTTCTTTTCGGCTCAGTCCCTTTATTAATCAGGGGTTGAATGAACTTGTCAACTTATCATCATGT
GTTTTATGCAGCGGATGCCCTTCCAGGTGCAACCCATTACTGGGAAACACCCCCTTGCCAACCAGCTTAG
CATTAAAATCCACACAATGTGTCCCTGTAAAAAAATCACATAAAAGGTTGAGAGAAATAACAAACCCAGC
AACCATCTGAGAAAGTACAAATGTAAAAAATGGTTATTTCATTGCTATTTGCATAAGTTTCCAAAAACTT
ACCAGGTCTGGAAAATGCCATGTCAAAGTCTTTCCAGGTTTTTCACAACCATAAGAACCCTGTCTGTAGT
CGTGTCATTCTACATCTGTGAGACTTGTTGCATAGTTGTGTGATTCTACATCTGTGAGACTTGTTGCAGT
GCTTGGCGAA