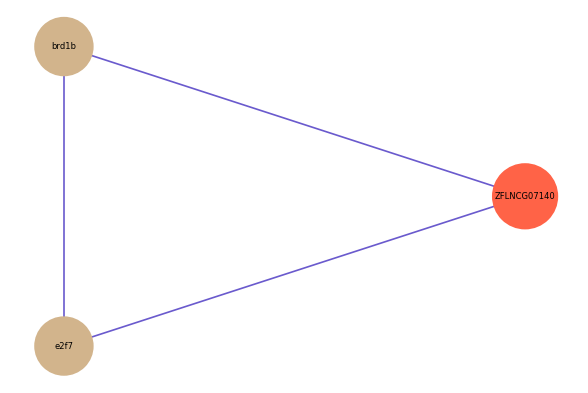
ZFLNCG07140
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800046 | sphere stage | 5azaCyD treatment | 1.46 |
| SRR514028 | retina | control morpholino | 0.94 |
| SRR594771 | endothelium | normal | 0.84 |
| SRR800049 | sphere stage | control treatment | 0.77 |
| SRR800043 | sphere stage | normal | 0.71 |
| SRR594769 | blood | normal | 0.54 |
| SRR658537 | bud | Gata6 morphant | 0.51 |
| SRR726542 | 5 dpf | infection with control | 0.34 |
| SRR372796 | bud | normal | 0.33 |
| SRR700534 | heart | control morpholino | 0.23 |
Express in tissues
Gene Ontology
Download
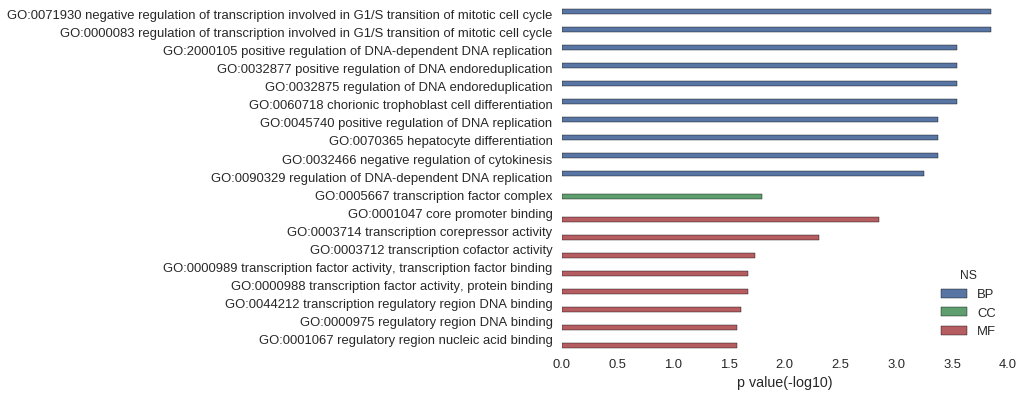
| GO | P value |
|---|---|
| GO:0071930 | 1.42e-04 |
| GO:0000083 | 1.42e-04 |
| GO:2000105 | 2.84e-04 |
| GO:0032877 | 2.84e-04 |
| GO:0032875 | 2.84e-04 |
| GO:0060718 | 2.84e-04 |
| GO:0045740 | 4.26e-04 |
| GO:0070365 | 4.26e-04 |
| GO:0032466 | 4.26e-04 |
| GO:0090329 | 5.68e-04 |
| GO:0005667 | 1.61e-02 |
| GO:0001047 | 1.42e-03 |
| GO:0003714 | 4.97e-03 |
| GO:0003712 | 1.87e-02 |
| GO:0000989 | 2.12e-02 |
| GO:0000988 | 2.15e-02 |
| GO:0044212 | 2.49e-02 |
| GO:0000975 | 2.71e-02 |
| GO:0001067 | 2.71e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07140
AATTTAAAAGTGCTGGCTAATGCATAACACAAATTCAGTAAGATAGTTATTCACATCCACCTTAATAAAA
TCTGATTATAATAAAGGCGTTTGAACAAAGACAATTTCAGACACCTTATTATTCTTTGGTCCCCTGCTCT
CTTACTGTGCTGACAAGAACTATAGCAGATGCCCAGTGGCTGGATGTGACAATGGAAACGGCAGAAAAAC
AAGATGAGTGGTATGTATAATGACGCGTTAAACTTGAGGAAGCACTGTGACTTGTTGGTTAATGGGAAAA
AATGAAAAATTGTAGAAAGTTGATTCTTATTTGACTGCAAGAGGATTGTTCTGGTGCAATCACCCTCAGC
ATTTTCCTACCTCATCATGGCTTTCTTGTCTATCTCAAGAGTTTGATTAAGATTGCAGTGCTATGGCGCA
CTTTGATAAGTAAGTAATGCATCTATGAAGCGTGCTTCACACAGATGTGTGGACTTGACAAACCACTCGT
AGAAACCCAAAAAGGCACTCTAGCACTAAATTCTCTGTGCACAAAAAGCTCATTTCTATATAGAGCACTT
CTTAGCCCCTTTCACGCAGAAAATTTACAGAACGACTTTTTC
AATTTAAAAGTGCTGGCTAATGCATAACACAAATTCAGTAAGATAGTTATTCACATCCACCTTAATAAAA
TCTGATTATAATAAAGGCGTTTGAACAAAGACAATTTCAGACACCTTATTATTCTTTGGTCCCCTGCTCT
CTTACTGTGCTGACAAGAACTATAGCAGATGCCCAGTGGCTGGATGTGACAATGGAAACGGCAGAAAAAC
AAGATGAGTGGTATGTATAATGACGCGTTAAACTTGAGGAAGCACTGTGACTTGTTGGTTAATGGGAAAA
AATGAAAAATTGTAGAAAGTTGATTCTTATTTGACTGCAAGAGGATTGTTCTGGTGCAATCACCCTCAGC
ATTTTCCTACCTCATCATGGCTTTCTTGTCTATCTCAAGAGTTTGATTAAGATTGCAGTGCTATGGCGCA
CTTTGATAAGTAAGTAATGCATCTATGAAGCGTGCTTCACACAGATGTGTGGACTTGACAAACCACTCGT
AGAAACCCAAAAAGGCACTCTAGCACTAAATTCTCTGTGCACAAAAAGCTCATTTCTATATAGAGCACTT
CTTAGCCCCTTTCACGCAGAAAATTTACAGAACGACTTTTTC