ZFLNCG07202
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647680 | head kidney | SVCV treatment | 28.25 |
| SRR516122 | skin | male and 3.5 year | 26.78 |
| SRR1647684 | spleen | SVCV treatment | 23.01 |
| SRR1647681 | head kidney | SVCV treatment | 21.06 |
| SRR1647683 | spleen | SVCV treatment | 20.75 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 20.70 |
| SRR516123 | skin | male and 3.5 year | 20.70 |
| SRR516124 | skin | male and 3.5 year | 20.45 |
| SRR891504 | liver | normal | 19.32 |
| SRR516128 | skin | male and 5 month | 17.89 |
Express in tissues
Correlated coding gene
Download
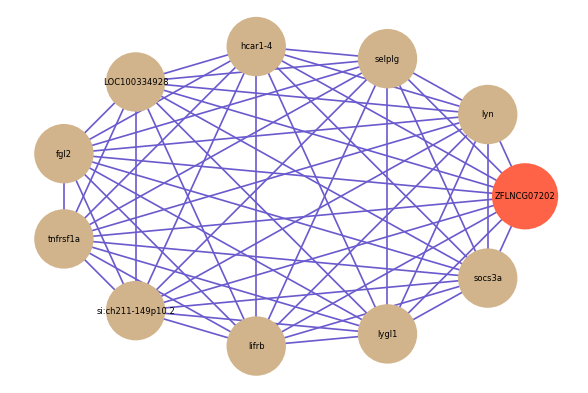
| gene | correlation coefficent |
|---|---|
| lyn | 0.60 |
| selplg | 0.60 |
| hcar1-4 | 0.59 |
| LOC100334928 | 0.59 |
| fgl2 | 0.58 |
| tnfrsf1a | 0.58 |
| si:ch211-149p10.2 | 0.58 |
| lifrb | 0.58 |
| lygl1 | 0.57 |
| socs3a | 0.57 |
Gene Ontology
Download
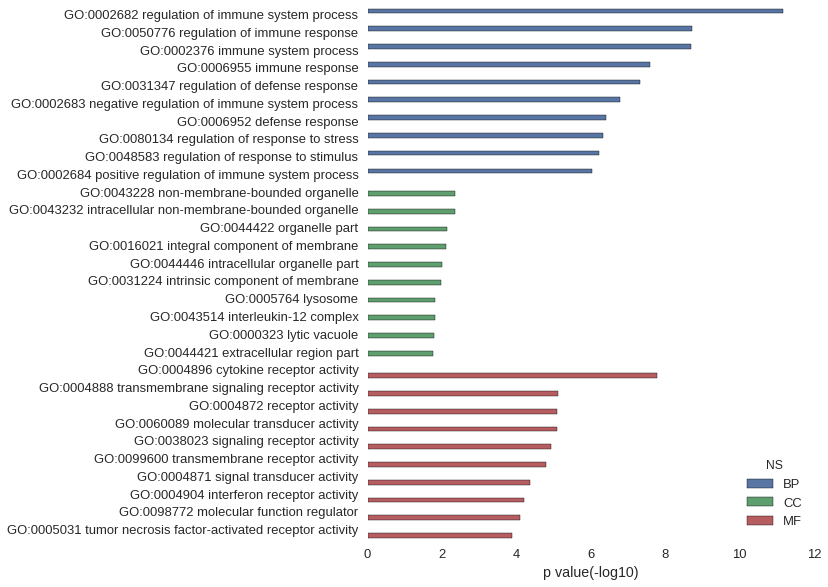
| GO | P value |
|---|---|
| GO:0002682 | 6.98e-12 |
| GO:0050776 | 1.88e-09 |
| GO:0002376 | 1.96e-09 |
| GO:0006955 | 2.52e-08 |
| GO:0031347 | 4.73e-08 |
| GO:0002683 | 1.60e-07 |
| GO:0006952 | 3.91e-07 |
| GO:0080134 | 4.61e-07 |
| GO:0048583 | 5.93e-07 |
| GO:0002684 | 9.25e-07 |
| GO:0043228 | 4.43e-03 |
| GO:0043232 | 4.43e-03 |
| GO:0044422 | 7.23e-03 |
| GO:0016021 | 7.73e-03 |
| GO:0044446 | 9.91e-03 |
| GO:0031224 | 1.05e-02 |
| GO:0005764 | 1.53e-02 |
| GO:0043514 | 1.57e-02 |
| GO:0000323 | 1.65e-02 |
| GO:0044421 | 1.69e-02 |
| GO:0004896 | 1.62e-08 |
| GO:0004888 | 7.74e-06 |
| GO:0004872 | 8.02e-06 |
| GO:0060089 | 8.02e-06 |
| GO:0038023 | 1.17e-05 |
| GO:0099600 | 1.57e-05 |
| GO:0004871 | 4.33e-05 |
| GO:0004904 | 6.17e-05 |
| GO:0098772 | 8.20e-05 |
| GO:0005031 | 1.29e-04 |
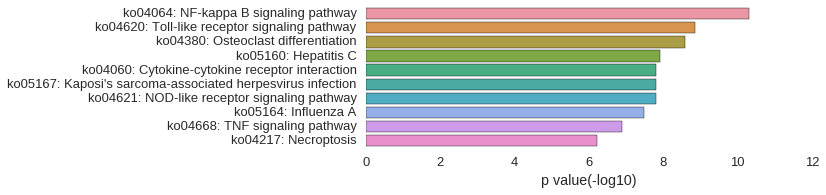
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07202
TCTGATCCAAGCGTCTCCCCGGAGAGTCGTCAGTCTATAGCAATCGATGATTGGCTAAACAAGTGACATG
TCTTGTGTGTTCTATAGTCTTTAGATCCGCTCACTTTAAAATCTCTGGTGTCAACACCTATTTGCTGACA
AAATAAGCTGACTTTTATTATATAGACAAACAAAATGTCAAAATTATGGGAAGCTTTAGAAAGATGGATG
AGTTTTCAGTTCTAC
TCTGATCCAAGCGTCTCCCCGGAGAGTCGTCAGTCTATAGCAATCGATGATTGGCTAAACAAGTGACATG
TCTTGTGTGTTCTATAGTCTTTAGATCCGCTCACTTTAAAATCTCTGGTGTCAACACCTATTTGCTGACA
AAATAAGCTGACTTTTATTATATAGACAAACAAAATGTCAAAATTATGGGAAGCTTTAGAAAGATGGATG
AGTTTTCAGTTCTAC