ZFLNCG07361
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR516129 | skin | male and 5 month | 62.31 |
| SRR516123 | skin | male and 3.5 year | 61.85 |
| SRR516133 | skin | male and 5 month | 60.29 |
| SRR516135 | skin | male and 5 month | 54.48 |
| SRR516125 | skin | male and 3.5 year | 45.08 |
| SRR941753 | posterior pectoral fin | normal | 40.63 |
| SRR941749 | anterior pectoral fin | normal | 40.02 |
| SRR516124 | skin | male and 3.5 year | 39.66 |
| SRR516132 | skin | male and 5 month | 39.37 |
| SRR516121 | skin | male and 3.5 year | 38.43 |
Express in tissues
Correlated coding gene
Download
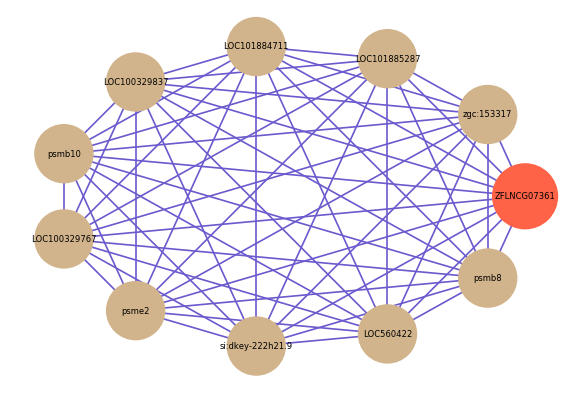
| gene | correlation coefficent |
|---|---|
| zgc:153317 | 0.65 |
| LOC101885287 | 0.65 |
| LOC101884711 | 0.64 |
| LOC100329837 | 0.64 |
| psmb10 | 0.63 |
| LOC100329767 | 0.63 |
| psme2 | 0.63 |
| si:dkey-222h21.9 | 0.63 |
| LOC560422 | 0.63 |
| psmb8 | 0.63 |
Gene Ontology
Download
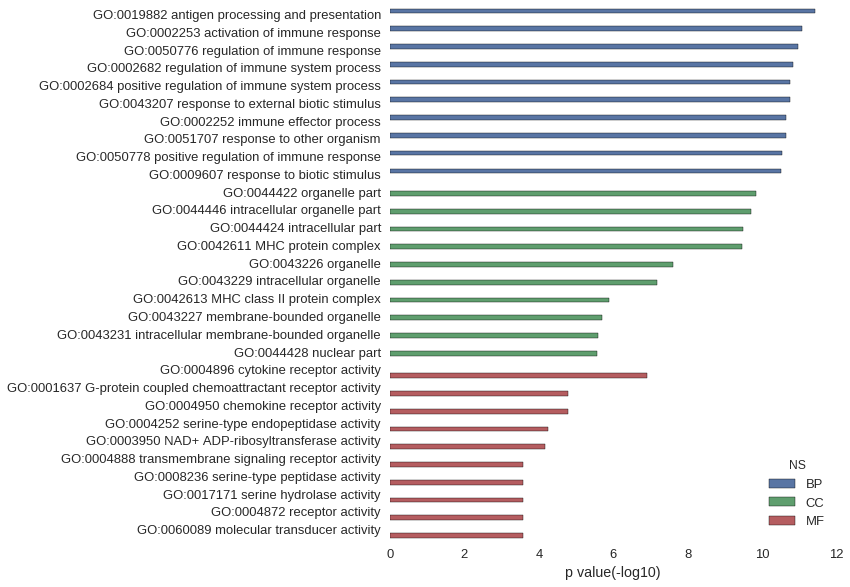
| GO | P value |
|---|---|
| GO:0019882 | 3.77e-12 |
| GO:0002253 | 8.69e-12 |
| GO:0050776 | 1.10e-11 |
| GO:0002682 | 1.46e-11 |
| GO:0002684 | 1.76e-11 |
| GO:0043207 | 1.81e-11 |
| GO:0002252 | 2.21e-11 |
| GO:0051707 | 2.34e-11 |
| GO:0050778 | 2.98e-11 |
| GO:0009607 | 3.16e-11 |
| GO:0044422 | 1.45e-10 |
| GO:0044446 | 2.02e-10 |
| GO:0044424 | 3.20e-10 |
| GO:0042611 | 3.49e-10 |
| GO:0043226 | 2.53e-08 |
| GO:0043229 | 6.57e-08 |
| GO:0042613 | 1.31e-06 |
| GO:0043227 | 1.96e-06 |
| GO:0043231 | 2.61e-06 |
| GO:0044428 | 2.68e-06 |
| GO:0004896 | 1.26e-07 |
| GO:0001637 | 1.67e-05 |
| GO:0004950 | 1.67e-05 |
| GO:0004252 | 5.81e-05 |
| GO:0003950 | 6.70e-05 |
| GO:0004888 | 2.60e-04 |
| GO:0008236 | 2.61e-04 |
| GO:0017171 | 2.61e-04 |
| GO:0004872 | 2.62e-04 |
| GO:0060089 | 2.62e-04 |
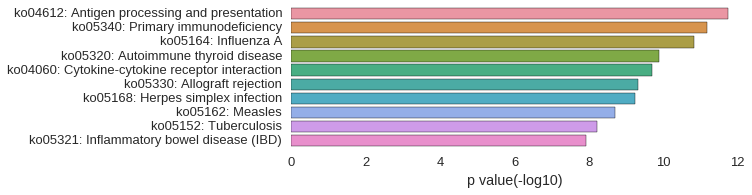
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07361
TTTCACCTCCCAATCCTGCCCCCTTGATCCTCAGCTTTTTGGTACAGTATGTAACACCCCCAAGTTGCCA
ACTTTGGTATAGTATCTGATGGCAGCGGCCGCCTGAAACCCCCGTAGTACAGTGACTAACCCAGTTCTCA
ACAGTGTTATTCAGACACCTCACTCCCGAACACCCTCAGACTTCTGGATTTTAGAGACAATGTTGGCAGG
TATCCACTATGCTGATACACAGGCATTTCTAGCTCCT
TTTCACCTCCCAATCCTGCCCCCTTGATCCTCAGCTTTTTGGTACAGTATGTAACACCCCCAAGTTGCCA
ACTTTGGTATAGTATCTGATGGCAGCGGCCGCCTGAAACCCCCGTAGTACAGTGACTAACCCAGTTCTCA
ACAGTGTTATTCAGACACCTCACTCCCGAACACCCTCAGACTTCTGGATTTTAGAGACAATGTTGGCAGG
TATCCACTATGCTGATACACAGGCATTTCTAGCTCCT