ZFLNCG07411
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR776507 | embryo | normal | 4.74 |
| ERR023144 | brain | normal | 4.60 |
| SRR776509 | embryo | Salmonella infected | 4.22 |
| ERR594417 | retina | normal | 4.13 |
| SRR519737 | 2 dpf | VD3 treatment | 3.97 |
| SRR1167757 | embryo | mpeg1 morpholino and bacterial infection | 3.88 |
| SRR1028003 | head | normal | 3.51 |
| SRR592703 | pineal gland | normal | 3.27 |
| SRR1028004 | head | normal | 3.01 |
| SRR519717 | 2 dpf | FETOH treatment | 2.97 |
Express in tissues
Correlated coding gene
Download
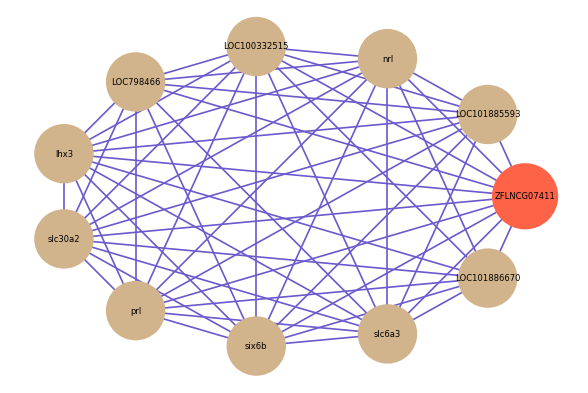
| gene | correlation coefficent |
|---|---|
| LOC101885593 | 0.62 |
| LOC101886670 | 0.57 |
| nrl | 0.57 |
| LOC100332515 | 0.56 |
| LOC798466 | 0.56 |
| lhx3 | 0.56 |
| slc30a2 | 0.56 |
| prl | 0.55 |
| six6b | 0.55 |
| slc6a3 | 0.54 |
Gene Ontology
Download
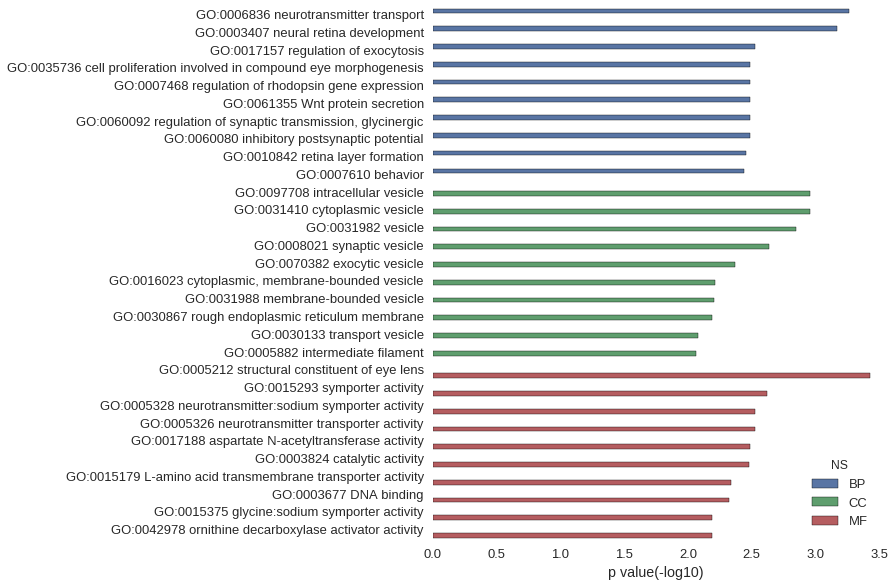
| GO | P value |
|---|---|
| GO:0006836 | 5.42e-04 |
| GO:0003407 | 6.76e-04 |
| GO:0017157 | 2.99e-03 |
| GO:0035736 | 3.27e-03 |
| GO:0007468 | 3.27e-03 |
| GO:0061355 | 3.27e-03 |
| GO:0060092 | 3.27e-03 |
| GO:0060080 | 3.27e-03 |
| GO:0010842 | 3.48e-03 |
| GO:0007610 | 3.67e-03 |
| GO:0097708 | 1.11e-03 |
| GO:0031410 | 1.11e-03 |
| GO:0031982 | 1.43e-03 |
| GO:0008021 | 2.32e-03 |
| GO:0070382 | 4.29e-03 |
| GO:0016023 | 6.10e-03 |
| GO:0031988 | 6.25e-03 |
| GO:0030867 | 6.53e-03 |
| GO:0030133 | 8.28e-03 |
| GO:0005882 | 8.67e-03 |
| GO:0005212 | 3.71e-04 |
| GO:0015293 | 2.42e-03 |
| GO:0005328 | 2.99e-03 |
| GO:0005326 | 2.99e-03 |
| GO:0017188 | 3.27e-03 |
| GO:0003824 | 3.30e-03 |
| GO:0015179 | 4.58e-03 |
| GO:0003677 | 4.80e-03 |
| GO:0015375 | 6.53e-03 |
| GO:0042978 | 6.53e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07411
TGTGTGTCAGCAGATAAACACTGATGTAGACACTGGAGCTGCCGTCATGTTATAGAGTCGGGGGCTGCAG
CTCTGCTCCTGGAGATCCGCCTACCTGCAGAGTCCAGCCCTGATCAAACACACACACACACACACACCTG
TCTGTAATCATCAGGTGCTGCTTTAGCTCCTGTTAGCTGCTTCAGGTGGGTTTGATCAGGGTTGTAGCTG
ATCTCTGCAGGAAGGTGAATGTCCAGGAGCAGGGTTGCTGCCCCCGACTCTGTCCCCCGTTGTAACGTGC
AGAACTCTGTCTTCAGCTCTCTGTGGGCTGGTTTGACTCCGGGTCTTCTCTGAACACACTCACCTGAGCT
GGCTGCGGTTCAGCAGGGGCAGGTAGTCAGGGATGGAGATGTAGATGAAGCTCTGGGCCTCCTGGATCAC
TCTGCTGATGGCCTCCAGGTCTTTGGTCCGCTCTCGAGTGCAGAAAACATCTGGAGAAGTCTAGAAAACA
CGAACATGCTCGGCTTCATCACTGAGCAGAATATTAACGCC
TGTGTGTCAGCAGATAAACACTGATGTAGACACTGGAGCTGCCGTCATGTTATAGAGTCGGGGGCTGCAG
CTCTGCTCCTGGAGATCCGCCTACCTGCAGAGTCCAGCCCTGATCAAACACACACACACACACACACCTG
TCTGTAATCATCAGGTGCTGCTTTAGCTCCTGTTAGCTGCTTCAGGTGGGTTTGATCAGGGTTGTAGCTG
ATCTCTGCAGGAAGGTGAATGTCCAGGAGCAGGGTTGCTGCCCCCGACTCTGTCCCCCGTTGTAACGTGC
AGAACTCTGTCTTCAGCTCTCTGTGGGCTGGTTTGACTCCGGGTCTTCTCTGAACACACTCACCTGAGCT
GGCTGCGGTTCAGCAGGGGCAGGTAGTCAGGGATGGAGATGTAGATGAAGCTCTGGGCCTCCTGGATCAC
TCTGCTGATGGCCTCCAGGTCTTTGGTCCGCTCTCGAGTGCAGAAAACATCTGGAGAAGTCTAGAAAACA
CGAACATGCTCGGCTTCATCACTGAGCAGAATATTAACGCC