ZFLNCG07453
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1028003 | head | normal | 45.36 |
| SRR1028002 | head | normal | 25.43 |
| SRR527832 | 16-36 hpf | normal | 25.38 |
| SRR592701 | pineal gland | normal | 22.07 |
| SRR1048061 | pineal gland | dark | 20.14 |
| SRR1035978 | 13 hpf | rx3-/- | 18.56 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 18.15 |
| SRR658543 | 6 somite | Gata5 morphan | 17.39 |
| SRR527834 | head | normal | 14.69 |
| SRR594771 | endothelium | normal | 14.62 |
Express in tissues
Correlated coding gene
Download
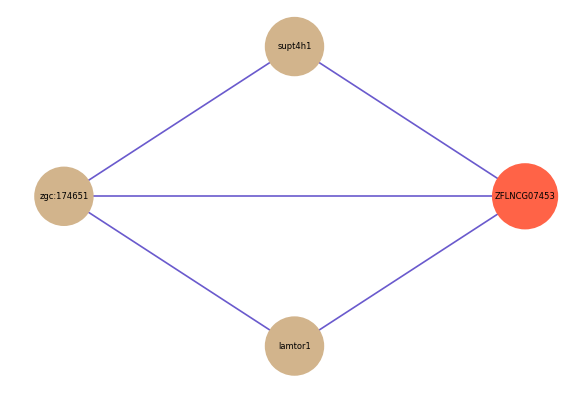
| gene | correlation coefficent |
|---|---|
| supt4h1 | 0.61 |
| lamtor1 | 0.57 |
| zgc:174651 | 0.51 |
Gene Ontology
Download
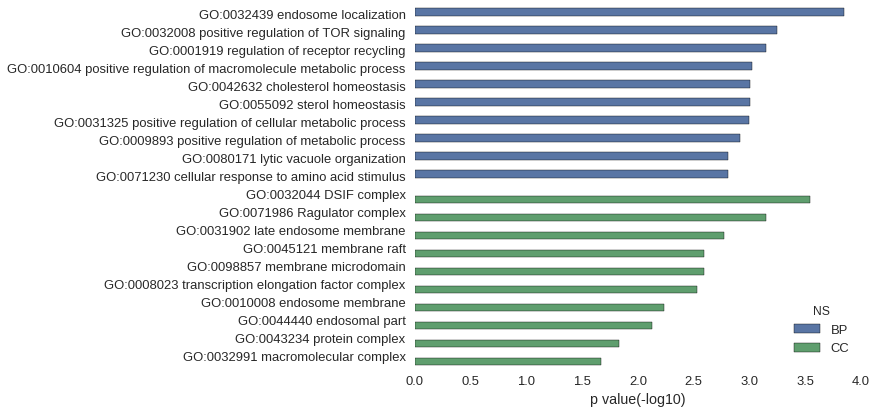
| GO | P value |
|---|---|
| GO:0032439 | 1.42e-04 |
| GO:0032008 | 5.68e-04 |
| GO:0001919 | 7.11e-04 |
| GO:0010604 | 9.45e-04 |
| GO:0042632 | 9.95e-04 |
| GO:0055092 | 9.95e-04 |
| GO:0031325 | 1.01e-03 |
| GO:0009893 | 1.22e-03 |
| GO:0080171 | 1.56e-03 |
| GO:0071230 | 1.56e-03 |
| GO:0032044 | 2.84e-04 |
| GO:0071986 | 7.11e-04 |
| GO:0031902 | 1.70e-03 |
| GO:0045121 | 2.56e-03 |
| GO:0098857 | 2.56e-03 |
| GO:0008023 | 2.98e-03 |
| GO:0010008 | 5.82e-03 |
| GO:0044440 | 7.52e-03 |
| GO:0043234 | 1.47e-02 |
| GO:0032991 | 2.14e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07453
AAATACATTTCATAGTACATGCTGCAAAATAGACCAGAATTAGGAAGGTAAATCGATTTATTAACTTCTT
ACCTGTCTCCGGTAATTTTCCGCAGTTAAATCCAGAGTTCGGTGTCGATCAGTCTTGACTCCTGCTCAAA
CTGAATAAACTTTGCCCGCTTTTCCTTCTTCTTCTCCTCCTCGCAGGCTAACACTCTTGCTAACGGCGGC
TAAAATATCCTCGCTCGCGCCGTCAGGAGCGATCTCCGATCCCGCGTCCTCTCGCCGCTATCTCTGGGCG
AGGAGTTAAAGTGTTACCGCAGATGGCAGGTGTGGATATCCGCTGTTCCAGATTTCAGATAAAAGTTATC
TCCATCCTCTCCCGTTCAGC
AAATACATTTCATAGTACATGCTGCAAAATAGACCAGAATTAGGAAGGTAAATCGATTTATTAACTTCTT
ACCTGTCTCCGGTAATTTTCCGCAGTTAAATCCAGAGTTCGGTGTCGATCAGTCTTGACTCCTGCTCAAA
CTGAATAAACTTTGCCCGCTTTTCCTTCTTCTTCTCCTCCTCGCAGGCTAACACTCTTGCTAACGGCGGC
TAAAATATCCTCGCTCGCGCCGTCAGGAGCGATCTCCGATCCCGCGTCCTCTCGCCGCTATCTCTGGGCG
AGGAGTTAAAGTGTTACCGCAGATGGCAGGTGTGGATATCCGCTGTTCCAGATTTCAGATAAAAGTTATC
TCCATCCTCTCCCGTTCAGC

