ZFLNCG07577
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562530 | liver | normal | 95.65 |
| SRR891504 | liver | normal | 66.33 |
| SRR1562529 | intestine and pancreas | normal | 12.48 |
| SRR1035239 | liver | transgenic mCherry control | 12.39 |
| SRR1562532 | spleen | normal | 8.53 |
| SRR1647682 | spleen | normal | 7.69 |
| SRR1647681 | head kidney | SVCV treatment | 6.75 |
| SRR1049952 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with gallein | 6.28 |
| ERR023146 | head kidney | normal | 5.59 |
| SRR1039571 | gastrointestinal | diet 0.1 NPM | 4.62 |
Express in tissues
Correlated coding gene
Download
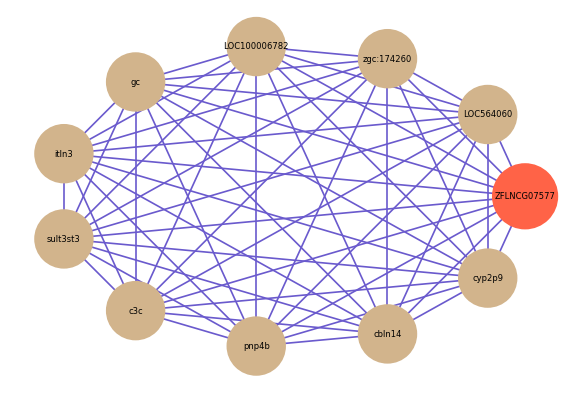
| gene | correlation coefficent |
|---|---|
| LOC564060 | 0.72 |
| zgc:174260 | 0.72 |
| LOC100006782 | 0.72 |
| gc | 0.72 |
| itln3 | 0.71 |
| sult3st3 | 0.70 |
| c3c | 0.70 |
| pnp4b | 0.70 |
| cbln14 | 0.70 |
| cyp2p9 | 0.69 |
Gene Ontology
Download
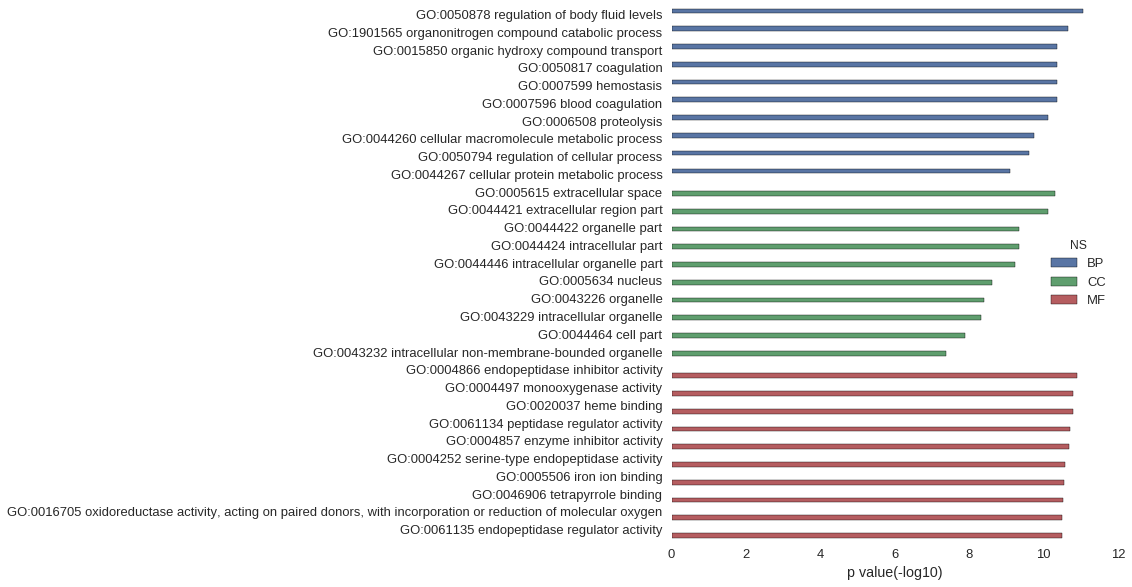
| GO | P value |
|---|---|
| GO:0050878 | 8.58e-12 |
| GO:1901565 | 2.24e-11 |
| GO:0015850 | 4.23e-11 |
| GO:0050817 | 4.31e-11 |
| GO:0007599 | 4.31e-11 |
| GO:0007596 | 4.31e-11 |
| GO:0006508 | 7.39e-11 |
| GO:0044260 | 1.79e-10 |
| GO:0050794 | 2.40e-10 |
| GO:0044267 | 7.87e-10 |
| GO:0005615 | 4.79e-11 |
| GO:0044421 | 7.61e-11 |
| GO:0044422 | 4.49e-10 |
| GO:0044424 | 4.51e-10 |
| GO:0044446 | 5.68e-10 |
| GO:0005634 | 2.50e-09 |
| GO:0043226 | 3.93e-09 |
| GO:0043229 | 4.70e-09 |
| GO:0044464 | 1.33e-08 |
| GO:0043232 | 4.29e-08 |
| GO:0004866 | 1.24e-11 |
| GO:0004497 | 1.63e-11 |
| GO:0020037 | 1.63e-11 |
| GO:0061134 | 2.00e-11 |
| GO:0004857 | 2.05e-11 |
| GO:0004252 | 2.63e-11 |
| GO:0005506 | 2.74e-11 |
| GO:0046906 | 3.08e-11 |
| GO:0016705 | 3.13e-11 |
| GO:0061135 | 3.27e-11 |
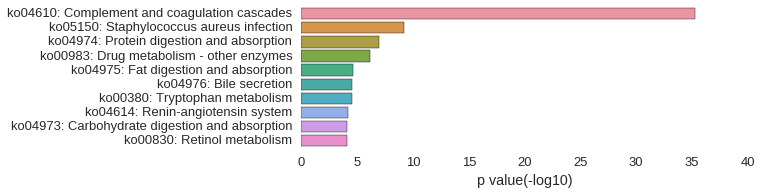
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07577
TAAACATAAGGTGAATAAATAGCGAACCCCTGCCGAATAAACCTAGCCTTGGTGGGACTGAAAAAAAGTC
CCGCACAGACATGTAGGCTATAGGTTCATGTCCCTATGGGGTCCTGTGGTCTAGATGATCTGACAAGCCA
CACCATCTCGTGGACATGTACTATAAAGACATCTTGTAGACAAATGTAGACAATTCCCTCAAGTGAATTT
GTCATCTGAAACATGCAGTGAAATGCTGTACCTGGAGCAACATATTTTTGCAAAAATCTAGGGTGAGGCA
CTTATTTCCAATGAGCCTGGCTAGATTTTTTGTTATAAAAAACAACAACAATTAAAAAATAAATATGGCT
CAATAGCAGTGAGACTGGTGTATAGTATACCGCATTGCACATAGGTTTACAGTAGATCTTATCCTTCACA
GATAAACACAGCAGTCCTCCATTCTCAGGCACTGCTGGAGCTGGACAGTTTTCTGGAAGTAACTTCTGAA
ACGCTGAAAAGAAGAAAGGACCTGTTCACTAGTATGACATGCAGTATAAGGATACATGCATGACATGTTA
TATTTTCATATTTCACATTATAAATTGCACTACAATATATTAAAGTTCTGGAAAATAACCAGAACATGAT
AGGTTATAACATTGTAAGTAATGTTACTATACCTTTGTTGTCTTGCCTCTGAATGATATCAGTTAATGCT
TGTAGTTGATTAAGAATGTTTCCGTGTTGGTATGACGTCCAGTTGTCAGATACAGTTTCCAAACAGTTTT
TTATGTTCTATAAAAATCATAAATTAATCTTAACGTTTTTTTTAATTTTTATTTTGTATCTGATGTGTGC
CTGCACTAGTCAACAAAACAGCAATACTATTTTATTAACGTTATCAAACAGATACTGTGTTTTAGCTGTA
GATAAAATGCTTTACCTGAAGCTGTTTGTCAAGCTGATCAAAAGAAAAGTCAGCGGGAAGGCATGAAGAA
TGAGATTGTCCATGAGTTAAAGTTGAGAGAAATGCTGTAGAAAAATCCCAAAGACATCTCCTGTAGTGCT
GTTCATAGAGACATTACAATCAAAGCATTCCAGCTGTGAAATAATCTAAATTATTCTTACCAAAGAGCAA
AAGCTGCTTCCATTTTCTGCCTATTGTTGCAGATTC
TAAACATAAGGTGAATAAATAGCGAACCCCTGCCGAATAAACCTAGCCTTGGTGGGACTGAAAAAAAGTC
CCGCACAGACATGTAGGCTATAGGTTCATGTCCCTATGGGGTCCTGTGGTCTAGATGATCTGACAAGCCA
CACCATCTCGTGGACATGTACTATAAAGACATCTTGTAGACAAATGTAGACAATTCCCTCAAGTGAATTT
GTCATCTGAAACATGCAGTGAAATGCTGTACCTGGAGCAACATATTTTTGCAAAAATCTAGGGTGAGGCA
CTTATTTCCAATGAGCCTGGCTAGATTTTTTGTTATAAAAAACAACAACAATTAAAAAATAAATATGGCT
CAATAGCAGTGAGACTGGTGTATAGTATACCGCATTGCACATAGGTTTACAGTAGATCTTATCCTTCACA
GATAAACACAGCAGTCCTCCATTCTCAGGCACTGCTGGAGCTGGACAGTTTTCTGGAAGTAACTTCTGAA
ACGCTGAAAAGAAGAAAGGACCTGTTCACTAGTATGACATGCAGTATAAGGATACATGCATGACATGTTA
TATTTTCATATTTCACATTATAAATTGCACTACAATATATTAAAGTTCTGGAAAATAACCAGAACATGAT
AGGTTATAACATTGTAAGTAATGTTACTATACCTTTGTTGTCTTGCCTCTGAATGATATCAGTTAATGCT
TGTAGTTGATTAAGAATGTTTCCGTGTTGGTATGACGTCCAGTTGTCAGATACAGTTTCCAAACAGTTTT
TTATGTTCTATAAAAATCATAAATTAATCTTAACGTTTTTTTTAATTTTTATTTTGTATCTGATGTGTGC
CTGCACTAGTCAACAAAACAGCAATACTATTTTATTAACGTTATCAAACAGATACTGTGTTTTAGCTGTA
GATAAAATGCTTTACCTGAAGCTGTTTGTCAAGCTGATCAAAAGAAAAGTCAGCGGGAAGGCATGAAGAA
TGAGATTGTCCATGAGTTAAAGTTGAGAGAAATGCTGTAGAAAAATCCCAAAGACATCTCCTGTAGTGCT
GTTCATAGAGACATTACAATCAAAGCATTCCAGCTGTGAAATAATCTAAATTATTCTTACCAAAGAGCAA
AAGCTGCTTCCATTTTCTGCCTATTGTTGCAGATTC