ZFLNCG07643
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR941753 | posterior pectoral fin | normal | 45.12 |
| SRR941749 | anterior pectoral fin | normal | 39.39 |
| SRR516123 | skin | male and 3.5 year | 19.99 |
| SRR516124 | skin | male and 3.5 year | 17.72 |
| SRR516126 | skin | male and 5 month | 17.45 |
| SRR516129 | skin | male and 5 month | 16.41 |
| SRR516127 | skin | male and 5 month | 15.75 |
| SRR516122 | skin | male and 3.5 year | 14.43 |
| SRR516125 | skin | male and 3.5 year | 14.42 |
| SRR516121 | skin | male and 3.5 year | 13.41 |
Express in tissues
Correlated coding gene
Download
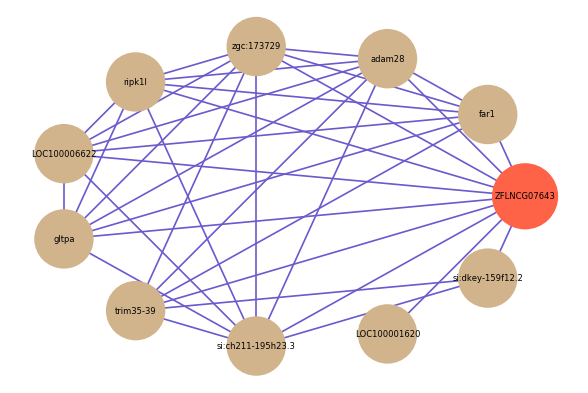
| gene | correlation coefficent |
|---|---|
| far1 | 0.60 |
| adam28 | 0.58 |
| zgc:173729 | 0.56 |
| ripk1l | 0.55 |
| LOC100001620 | 0.55 |
| LOC100006622 | 0.54 |
| si:ch211-195h23.3 | 0.53 |
| si:dkey-159f12.2 | 0.53 |
| gltpa | 0.53 |
| trim35-39 | 0.53 |
Gene Ontology
Download
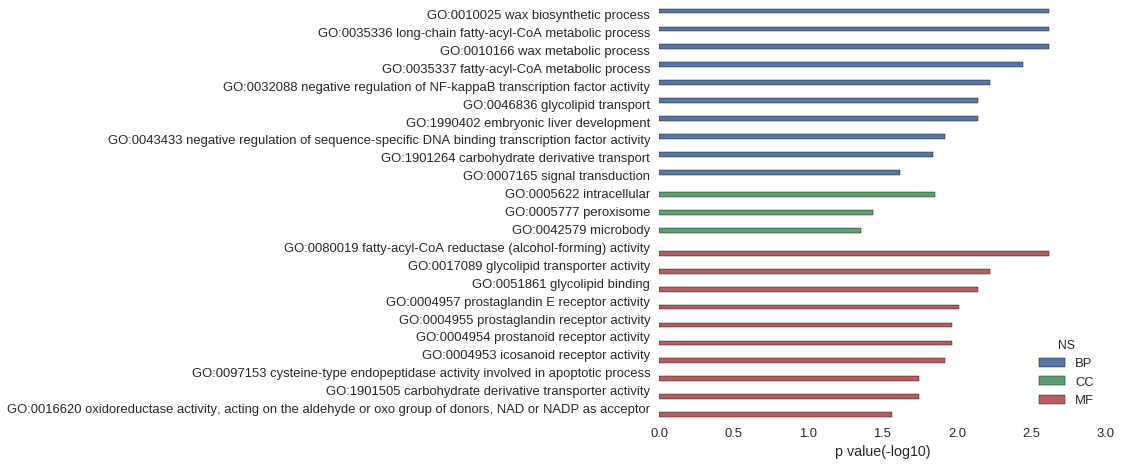
| GO | P value |
|---|---|
| GO:0010025 | 2.41e-03 |
| GO:0035336 | 2.41e-03 |
| GO:0010166 | 2.41e-03 |
| GO:0035337 | 3.62e-03 |
| GO:0032088 | 6.03e-03 |
| GO:0046836 | 7.23e-03 |
| GO:1990402 | 7.23e-03 |
| GO:0043433 | 1.20e-02 |
| GO:1901264 | 1.44e-02 |
| GO:0007165 | 2.41e-02 |
| GO:0005622 | 1.41e-02 |
| GO:0005777 | 3.68e-02 |
| GO:0042579 | 4.38e-02 |
| GO:0080019 | 2.41e-03 |
| GO:0017089 | 6.03e-03 |
| GO:0051861 | 7.23e-03 |
| GO:0004957 | 9.63e-03 |
| GO:0004955 | 1.08e-02 |
| GO:0004954 | 1.08e-02 |
| GO:0004953 | 1.20e-02 |
| GO:0097153 | 1.80e-02 |
| GO:1901505 | 1.80e-02 |
| GO:0016620 | 2.74e-02 |
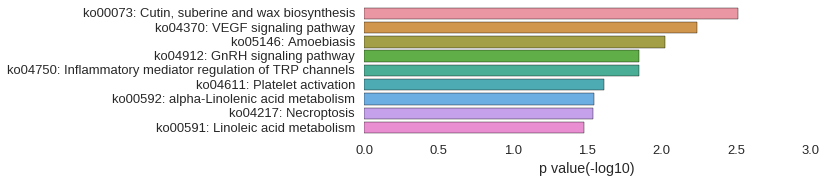
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07643
TAGACGTGTACACTTATGCTCAAACTATTTACATAGATTCTGAAGAGAAAGCTTTATGATAAGCAGCGTA
AGTTTAATTCTTTTTTCTTGTCTAACTCTTTTTGTCGATTCTGCACTTTAAACCTTAGTTTCTGTCATGG
AATTTTGTGGCTTTGTGTTTGCGCCCACTCGATTGTGTCATTCCATTCTATATGCCAGAGCTTGGATTTG
TAGGATAAGCCTTTCAGAAATATTTTCTGAAGTCAAACACTTAATATTGTGGCTTAAAATCCTCCGTAAT
GACTTCCTCTTTTATTTTTTTGTTCTATAATACTTTTCTGCTGCTGTTGAATGTGTTTTTTGTTCCTCAA
ATTCATAAAAGTCTTCATGTCTTAACTTGTGTTTATCTTGTGTAGGTTTTTGGGACTCGGAACTGTGCAA
TAAGAACAAGTCTTCTGTGTCTGGGGAAACTGCTTGTGATGGTTCAAAACAGAGACTAAAAAGAAGTCCA
GCCGTCCCGTTTACCCGGACCCCTTCTGAACTTGAATGCAGCCAATCCAACGGTGCACTTTGCTTCATCA
ATCCCCTCTTCCTGCAGACACACCAATCTAACCCTTCACCCAAAAAGCAAGAACCATCCCCAGGTGACGA
AGGTGGTGGCTCTCAGCACCACCACAG
TAGACGTGTACACTTATGCTCAAACTATTTACATAGATTCTGAAGAGAAAGCTTTATGATAAGCAGCGTA
AGTTTAATTCTTTTTTCTTGTCTAACTCTTTTTGTCGATTCTGCACTTTAAACCTTAGTTTCTGTCATGG
AATTTTGTGGCTTTGTGTTTGCGCCCACTCGATTGTGTCATTCCATTCTATATGCCAGAGCTTGGATTTG
TAGGATAAGCCTTTCAGAAATATTTTCTGAAGTCAAACACTTAATATTGTGGCTTAAAATCCTCCGTAAT
GACTTCCTCTTTTATTTTTTTGTTCTATAATACTTTTCTGCTGCTGTTGAATGTGTTTTTTGTTCCTCAA
ATTCATAAAAGTCTTCATGTCTTAACTTGTGTTTATCTTGTGTAGGTTTTTGGGACTCGGAACTGTGCAA
TAAGAACAAGTCTTCTGTGTCTGGGGAAACTGCTTGTGATGGTTCAAAACAGAGACTAAAAAGAAGTCCA
GCCGTCCCGTTTACCCGGACCCCTTCTGAACTTGAATGCAGCCAATCCAACGGTGCACTTTGCTTCATCA
ATCCCCTCTTCCTGCAGACACACCAATCTAACCCTTCACCCAAAAAGCAAGAACCATCCCCAGGTGACGA
AGGTGGTGGCTCTCAGCACCACCACAG