ZFLNCG07773
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 100.92 |
| SRR372793 | shield | normal | 81.91 |
| SRR1648856 | brain | normal | 78.31 |
| ERR023143 | swim bladder | normal | 59.67 |
| ERR023145 | heart | normal | 57.46 |
| SRR372802 | 5 dpf | normal | 52.77 |
| SRR372791 | dome | normal | 50.97 |
| SRR957181 | heart | normal | 48.48 |
| SRR1035237 | liver | transgenic UHRF1 | 44.51 |
| SRR1299129 | caudal fin | Seven days time after treatment | 42.37 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
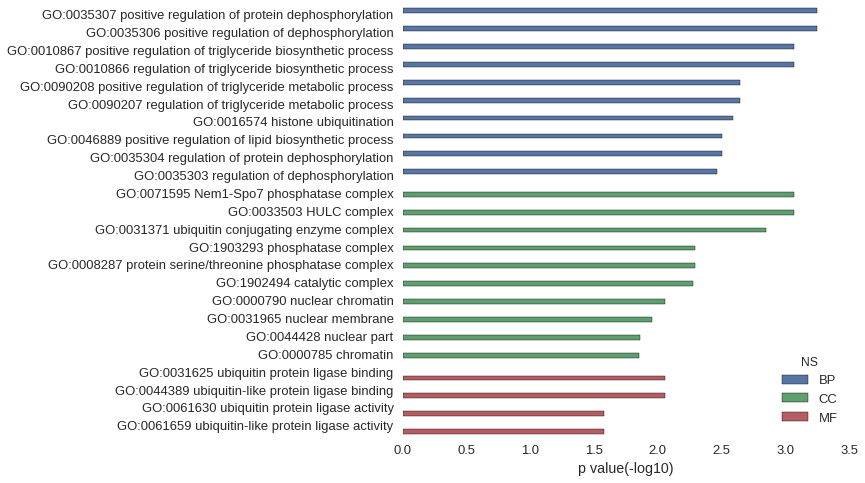
| GO | P value |
|---|---|
| GO:0035307 | 5.68e-04 |
| GO:0035306 | 5.68e-04 |
| GO:0010867 | 8.53e-04 |
| GO:0010866 | 8.53e-04 |
| GO:0090208 | 2.27e-03 |
| GO:0090207 | 2.27e-03 |
| GO:0016574 | 2.56e-03 |
| GO:0046889 | 3.12e-03 |
| GO:0035304 | 3.12e-03 |
| GO:0035303 | 3.41e-03 |
| GO:0071595 | 8.53e-04 |
| GO:0033503 | 8.53e-04 |
| GO:0031371 | 1.42e-03 |
| GO:1903293 | 5.11e-03 |
| GO:0008287 | 5.11e-03 |
| GO:1902494 | 5.34e-03 |
| GO:0000790 | 8.78e-03 |
| GO:0031965 | 1.10e-02 |
| GO:0044428 | 1.39e-02 |
| GO:0000785 | 1.41e-02 |
| GO:0031625 | 8.78e-03 |
| GO:0044389 | 8.78e-03 |
| GO:0061630 | 2.65e-02 |
| GO:0061659 | 2.65e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07773
AGAAAGCAAAGGTTTTAATTGGGACGGAGTGAAGGGAAGTAAAGAAAGTCAAAGGCAACAACAAACCTCA
TTAGAGAGATTCACTATAGCCCAGCAGAATCGAGCAACTGAGATCACATGACATTTCATCCCTAAAATGA
TACCCGATGAAAGCCGTCGTCTAAAAAGTGTCGCATAGTGACACCATGGCAGTGGATTCAACAGTAGATT
CCTCCTGACAGCTGAAAGAGCTACATCTGTATAAATAATGGGTATAAAGCAGTAAAGCAGAACAGTGATA
GGGTCAAAGCAGATGAATTACATACTTAGAAAAATTAAAGAGAAAGAGCAGAATCTTTTTACATTGTTTT
ACAAGCTTTCTTTAAAAAAT
AGAAAGCAAAGGTTTTAATTGGGACGGAGTGAAGGGAAGTAAAGAAAGTCAAAGGCAACAACAAACCTCA
TTAGAGAGATTCACTATAGCCCAGCAGAATCGAGCAACTGAGATCACATGACATTTCATCCCTAAAATGA
TACCCGATGAAAGCCGTCGTCTAAAAAGTGTCGCATAGTGACACCATGGCAGTGGATTCAACAGTAGATT
CCTCCTGACAGCTGAAAGAGCTACATCTGTATAAATAATGGGTATAAAGCAGTAAAGCAGAACAGTGATA
GGGTCAAAGCAGATGAATTACATACTTAGAAAAATTAAAGAGAAAGAGCAGAATCTTTTTACATTGTTTT
ACAAGCTTTCTTTAAAAAAT