ZFLNCG07909
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1048059 | pineal gland | light | 57.65 |
| SRR592703 | pineal gland | normal | 47.20 |
| SRR527832 | 16-36 hpf | normal | 37.81 |
| SRR516133 | skin | male and 5 month | 36.16 |
| SRR1048061 | pineal gland | dark | 25.04 |
| SRR700534 | heart | control morpholino | 23.91 |
| SRR065197 | 3 dpf | U1C knockout | 17.28 |
| SRR700537 | heart | Gata4 morpholino | 15.93 |
| SRR797914 | embryo | pbx2/4-MO | 15.90 |
| SRR658537 | bud | Gata6 morphant | 15.58 |
Express in tissues
Correlated coding gene
Download
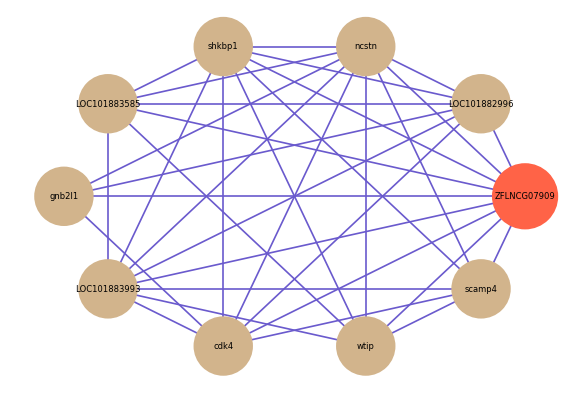
| gene | correlation coefficent |
|---|---|
| LOC101882996 | 0.54 |
| ncstn | 0.54 |
| shkbp1 | 0.52 |
| LOC101883585 | 0.52 |
| gnb2l1 | 0.52 |
| LOC101883993 | 0.52 |
| wtip | 0.51 |
| cdk4 | 0.51 |
| scamp4 | 0.51 |
Gene Ontology
Download
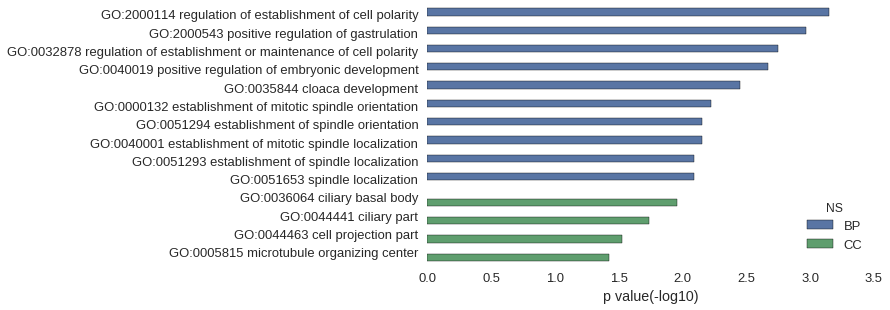
| GO | P value |
|---|---|
| GO:2000114 | 7.11e-04 |
| GO:2000543 | 1.07e-03 |
| GO:0032878 | 1.78e-03 |
| GO:0040019 | 2.13e-03 |
| GO:0035844 | 3.55e-03 |
| GO:0000132 | 6.03e-03 |
| GO:0051294 | 7.09e-03 |
| GO:0040001 | 7.09e-03 |
| GO:0051293 | 8.15e-03 |
| GO:0051653 | 8.15e-03 |
| GO:0036064 | 1.10e-02 |
| GO:0044441 | 1.83e-02 |
| GO:0044463 | 2.98e-02 |
| GO:0005815 | 3.78e-02 |
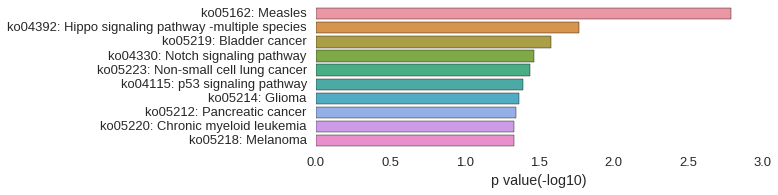
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG07909
TGAGAATGCGACTGTCGGGCGATGAGTGCAGTGTCCGCAGGTGATTTACCTGCGCGAACCGGCGGCGTGC
CCGCGCTCATGCAGTCATGCAACGTGCGGTAAAATCCTCGTGCCCGGTGATTAAAGTGCCTGAAGAAGCC
GGAGACCAGCAATGGAGGAGTAGCTGAAACAAACACCTGATCCGTGTGTGCCATGGGGAACTGCTGGGCT
CACTGTTTCGGGCTCTTTAGGAGAGAAGCCAACAGGATACAGAGAGGAGGAGGGTGAGCCATTCATGGGA
CTTGTGATGTGATTTTATTGAAAGGTTATCTGTGACGAAAACATCATTCGCCGGTTGGAGTAAAACTGGT
TTTATATTAGGGCATGTGGACTCTCTCCTTAATAATATGATATCAGAAAAGTGCTCTTTGCAAACTCTAA
ATTGGTAAATAACATTAGTAGCTAATGACTATCAAAATACAGAGTTCTCACAGTCAACTTGATCGTACGT
CCCCTCAATGATTCGCCGGCT
TGAGAATGCGACTGTCGGGCGATGAGTGCAGTGTCCGCAGGTGATTTACCTGCGCGAACCGGCGGCGTGC
CCGCGCTCATGCAGTCATGCAACGTGCGGTAAAATCCTCGTGCCCGGTGATTAAAGTGCCTGAAGAAGCC
GGAGACCAGCAATGGAGGAGTAGCTGAAACAAACACCTGATCCGTGTGTGCCATGGGGAACTGCTGGGCT
CACTGTTTCGGGCTCTTTAGGAGAGAAGCCAACAGGATACAGAGAGGAGGAGGGTGAGCCATTCATGGGA
CTTGTGATGTGATTTTATTGAAAGGTTATCTGTGACGAAAACATCATTCGCCGGTTGGAGTAAAACTGGT
TTTATATTAGGGCATGTGGACTCTCTCCTTAATAATATGATATCAGAAAAGTGCTCTTTGCAAACTCTAA
ATTGGTAAATAACATTAGTAGCTAATGACTATCAAAATACAGAGTTCTCACAGTCAACTTGATCGTACGT
CCCCTCAATGATTCGCCGGCT