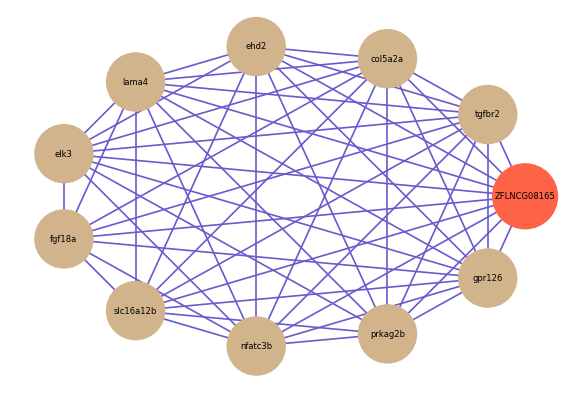
ZFLNCG08165
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR145631 | skeletal muscle | 32 degree_C to 16 degree_C | 127.53 |
| SRR516124 | skin | male and 3.5 year | 112.34 |
| ERR145635 | skeletal muscle | 27 degree_C to 16 degree_C | 110.81 |
| SRR891510 | muscle | normal | 90.42 |
| SRR1291417 | 5 dpi | injected marinum | 77.55 |
| ERR145646 | skeletal muscle | 32 degree_C to 27 degree_C | 65.56 |
| SRR516125 | skin | male and 3.5 year | 64.16 |
| SRR941749 | anterior pectoral fin | normal | 62.68 |
| SRR527833 | 5 dpf | normal | 62.12 |
| SRR527834 | head | normal | 59.12 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
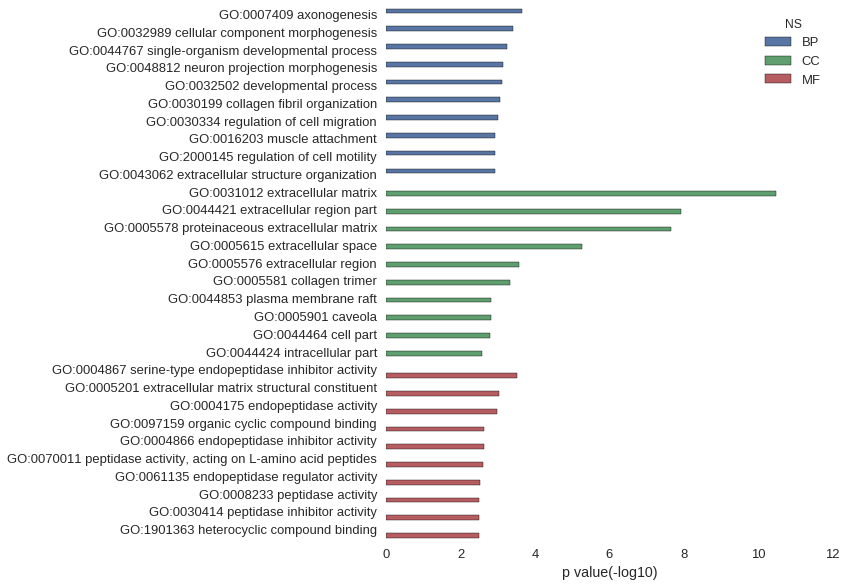
| GO | P value |
|---|---|
| GO:0007409 | 2.19e-04 |
| GO:0032989 | 4.01e-04 |
| GO:0044767 | 5.82e-04 |
| GO:0048812 | 7.38e-04 |
| GO:0032502 | 7.86e-04 |
| GO:0030199 | 8.88e-04 |
| GO:0030334 | 9.80e-04 |
| GO:0016203 | 1.18e-03 |
| GO:2000145 | 1.19e-03 |
| GO:0043062 | 1.21e-03 |
| GO:0031012 | 3.28e-11 |
| GO:0044421 | 1.17e-08 |
| GO:0005578 | 2.23e-08 |
| GO:0005615 | 5.37e-06 |
| GO:0005576 | 2.75e-04 |
| GO:0005581 | 4.71e-04 |
| GO:0044853 | 1.51e-03 |
| GO:0005901 | 1.51e-03 |
| GO:0044464 | 1.66e-03 |
| GO:0044424 | 2.61e-03 |
| GO:0004867 | 3.02e-04 |
| GO:0005201 | 9.02e-04 |
| GO:0004175 | 1.02e-03 |
| GO:0097159 | 2.34e-03 |
| GO:0004866 | 2.41e-03 |
| GO:0070011 | 2.45e-03 |
| GO:0061135 | 2.96e-03 |
| GO:0008233 | 3.20e-03 |
| GO:0030414 | 3.21e-03 |
| GO:1901363 | 3.24e-03 |
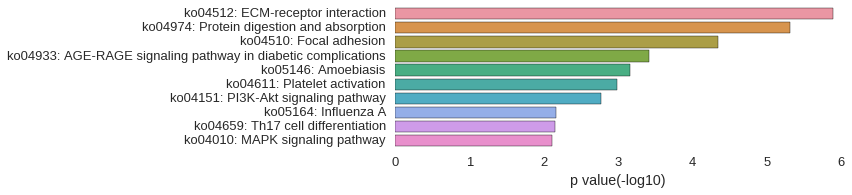
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08165
TTCACTTAGCCCTGGCGGCAGCCCTGGAAGATAAGGAAAGCCTCAGTACACTGTACATAATCTACATGAA
GCTTGCAGAGATTCATGCCAACTACATGCCAGATGCTGAACTATCTAAAAGCTACATGGACAGAGCGCAG
AGTCTGAGGAATGAACTGGCAGGATACACAGACTCAAAAGACACAGAAGAAACAGACGAGGCTGATGCTG
ACATTGACAACAAATCTGCTCATTCAGACGCCAGCAGTGGCACAAGCACCCTCACAGAGTCCAGCATGAT
CAATACCAGCCTTACAATCAATGCCCAGAAAGAGCCTGAGCACTCCAATGGAAGTCACAAAGACGATACA
AACACCAACAGATCAGAT
TTCACTTAGCCCTGGCGGCAGCCCTGGAAGATAAGGAAAGCCTCAGTACACTGTACATAATCTACATGAA
GCTTGCAGAGATTCATGCCAACTACATGCCAGATGCTGAACTATCTAAAAGCTACATGGACAGAGCGCAG
AGTCTGAGGAATGAACTGGCAGGATACACAGACTCAAAAGACACAGAAGAAACAGACGAGGCTGATGCTG
ACATTGACAACAAATCTGCTCATTCAGACGCCAGCAGTGGCACAAGCACCCTCACAGAGTCCAGCATGAT
CAATACCAGCCTTACAATCAATGCCCAGAAAGAGCCTGAGCACTCCAATGGAAGTCACAAAGACGATACA
AACACCAACAGATCAGAT