ZFLNCG08267
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372791 | dome | normal | 59.39 |
| SRR372793 | shield | normal | 55.01 |
| ERR023143 | swim bladder | normal | 54.38 |
| SRR1621206 | embryo | normal | 36.75 |
| SRR1562533 | testis | normal | 32.72 |
| SRR1021213 | sphere stage | normal | 31.97 |
| SRR1049814 | 256 cell | normal | 28.81 |
| SRR800043 | sphere stage | normal | 27.97 |
| SRR800046 | sphere stage | 5azaCyD treatment | 27.64 |
| SRR372789 | 1K cell | normal | 25.71 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
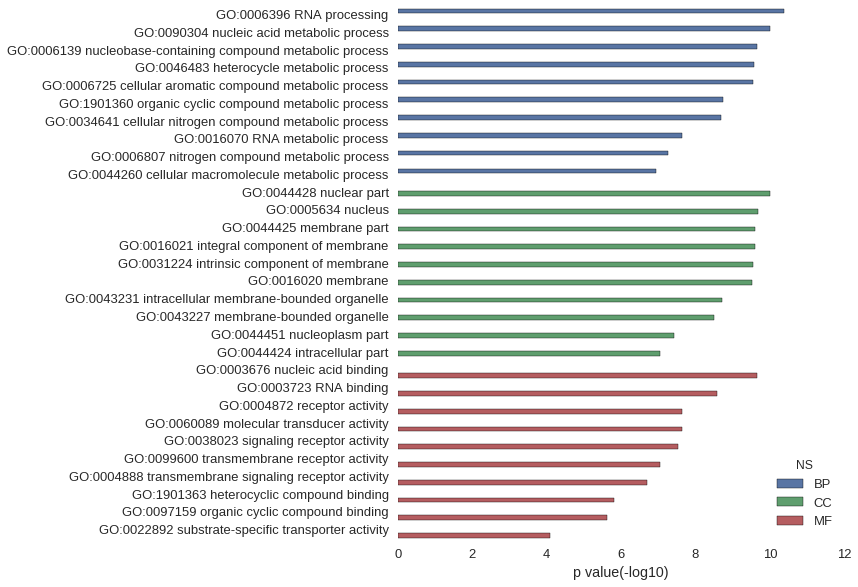
| GO | P value |
|---|---|
| GO:0006396 | 4.25e-11 |
| GO:0090304 | 9.72e-11 |
| GO:0006139 | 2.21e-10 |
| GO:0046483 | 2.70e-10 |
| GO:0006725 | 2.85e-10 |
| GO:1901360 | 1.77e-09 |
| GO:0034641 | 2.01e-09 |
| GO:0016070 | 2.36e-08 |
| GO:0006807 | 5.62e-08 |
| GO:0044260 | 1.12e-07 |
| GO:0044428 | 9.61e-11 |
| GO:0005634 | 2.03e-10 |
| GO:0044425 | 2.50e-10 |
| GO:0016021 | 2.54e-10 |
| GO:0031224 | 2.84e-10 |
| GO:0016020 | 2.92e-10 |
| GO:0043231 | 1.95e-09 |
| GO:0043227 | 3.25e-09 |
| GO:0044451 | 3.85e-08 |
| GO:0044424 | 8.99e-08 |
| GO:0003676 | 2.15e-10 |
| GO:0003723 | 2.59e-09 |
| GO:0004872 | 2.32e-08 |
| GO:0060089 | 2.32e-08 |
| GO:0038023 | 2.85e-08 |
| GO:0099600 | 9.15e-08 |
| GO:0004888 | 2.06e-07 |
| GO:1901363 | 1.55e-06 |
| GO:0097159 | 2.34e-06 |
| GO:0022892 | 8.09e-05 |
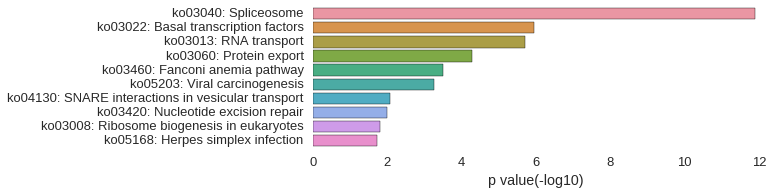
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08267
AATGACAGGTTTAAATACTCAGCAAGACTAAAGCTCTAAATATTAAAGGCATTTTCTGCATTGTAATCTT
GATACACAGGTTCAATGTTGCTGGGCAATGTTGCTTGGTTATTTTTCTATTAAGATTGGGCAACAGTATT
GTATCTAATGTAGCCCTAATTAGTTGCTCTGTGTCTCACCCGGTTGCCCTTTTATGGCATCATTGTTCAA
AACAACATTGCTGAGTAATATCTAATTTTTACATGTTAAAGAGACTGTTTACCCAAAAATATACAAGCTT
AAATATTACATCAGTATGAACTAATGATGAGTTAAGTAATGATGACTGACTAATTAAGAACTGAACCTAA
GGGGATAGTTCACCCCAAAACAAACACATTTAAACTCTCCTTTCATAATTCTAAATCGGTTCAATTTATT
TCTTCTGTTGGACACTAAAAAAGATTTTTTGAAGAATGTTCGAAACCGG
AATGACAGGTTTAAATACTCAGCAAGACTAAAGCTCTAAATATTAAAGGCATTTTCTGCATTGTAATCTT
GATACACAGGTTCAATGTTGCTGGGCAATGTTGCTTGGTTATTTTTCTATTAAGATTGGGCAACAGTATT
GTATCTAATGTAGCCCTAATTAGTTGCTCTGTGTCTCACCCGGTTGCCCTTTTATGGCATCATTGTTCAA
AACAACATTGCTGAGTAATATCTAATTTTTACATGTTAAAGAGACTGTTTACCCAAAAATATACAAGCTT
AAATATTACATCAGTATGAACTAATGATGAGTTAAGTAATGATGACTGACTAATTAAGAACTGAACCTAA
GGGGATAGTTCACCCCAAAACAAACACATTTAAACTCTCCTTTCATAATTCTAAATCGGTTCAATTTATT
TCTTCTGTTGGACACTAAAAAAGATTTTTTGAAGAATGTTCGAAACCGG