ZFLNCG08288
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1621206 | embryo | normal | 974.43 |
| SRR1049814 | 256 cell | normal | 942.34 |
| SRR372793 | shield | normal | 559.99 |
| SRR372796 | bud | normal | 402.21 |
| SRR372791 | dome | normal | 377.01 |
| SRR372787 | 2-4 cell | normal | 349.23 |
| SRR748488 | sperm | normal | 246.19 |
| SRR748490 | 1 kcell | normal | 214.90 |
| SRR658539 | bud | Gata5/6 morphant | 204.30 |
| SRR658533 | bud | normal | 193.95 |
Express in tissues
Correlated coding gene
Download
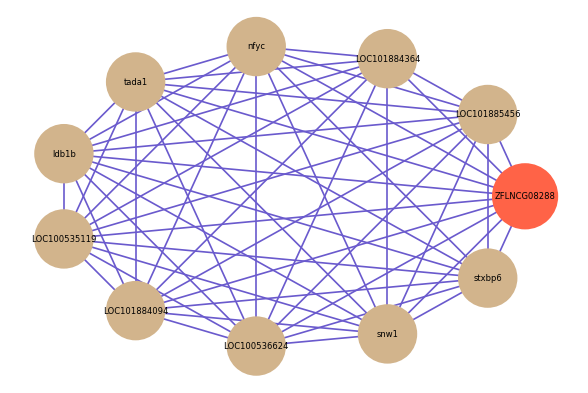
| gene | correlation coefficent |
|---|---|
| LOC101885456 | 0.80 |
| LOC101884364 | 0.72 |
| nfyc | 0.71 |
| tada1 | 0.70 |
| ldb1b | 0.70 |
| LOC100535119 | 0.68 |
| LOC101884094 | 0.67 |
| LOC100536624 | 0.67 |
| snw1 | 0.67 |
| stxbp6 | 0.65 |
Gene Ontology
Download
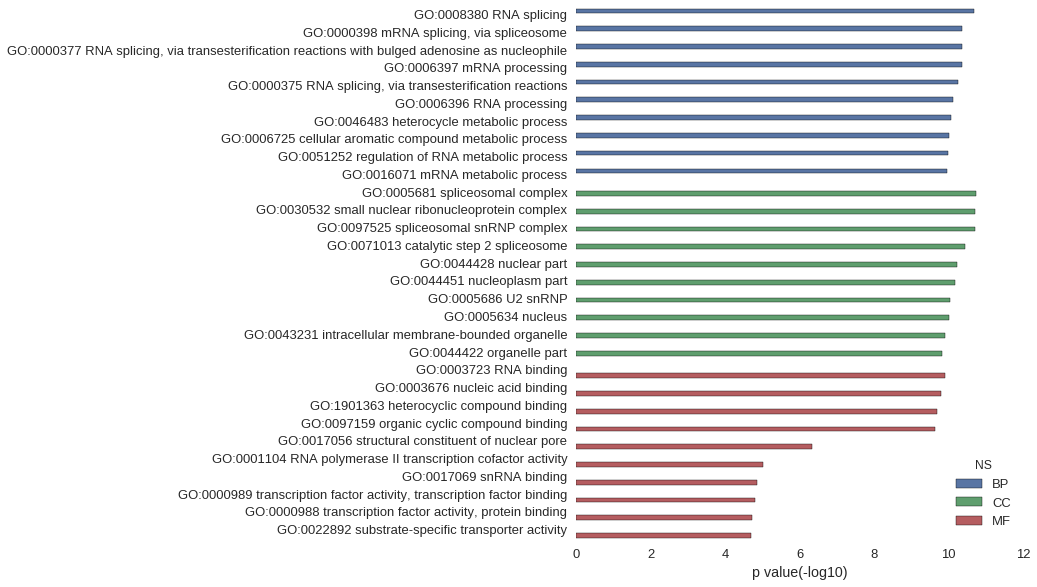
| GO | P value |
|---|---|
| GO:0008380 | 2.05e-11 |
| GO:0000398 | 4.35e-11 |
| GO:0000377 | 4.35e-11 |
| GO:0006397 | 4.42e-11 |
| GO:0000375 | 5.44e-11 |
| GO:0006396 | 7.56e-11 |
| GO:0046483 | 8.72e-11 |
| GO:0006725 | 9.82e-11 |
| GO:0051252 | 1.05e-10 |
| GO:0016071 | 1.06e-10 |
| GO:0005681 | 1.84e-11 |
| GO:0030532 | 1.91e-11 |
| GO:0097525 | 1.91e-11 |
| GO:0071013 | 3.49e-11 |
| GO:0044428 | 6.01e-11 |
| GO:0044451 | 6.56e-11 |
| GO:0005686 | 9.14e-11 |
| GO:0005634 | 9.52e-11 |
| GO:0043231 | 1.25e-10 |
| GO:0044422 | 1.46e-10 |
| GO:0003723 | 1.20e-10 |
| GO:0003676 | 1.57e-10 |
| GO:1901363 | 2.03e-10 |
| GO:0097159 | 2.28e-10 |
| GO:0017056 | 4.60e-07 |
| GO:0001104 | 9.95e-06 |
| GO:0017069 | 1.41e-05 |
| GO:0000989 | 1.63e-05 |
| GO:0000988 | 1.88e-05 |
| GO:0022892 | 2.04e-05 |
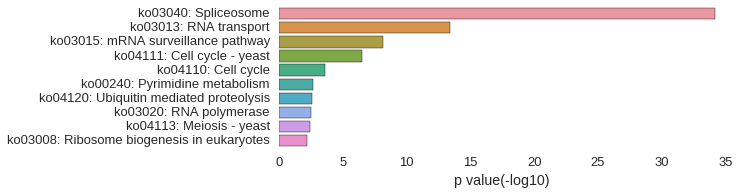
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08288
CCGTCGCCTTCATCGCTGTGTCTCCGGCGGGACATTTCTGCAGTTCAGCGAACACTGAGGAGGGAAACAC
TGAGGGAAATGCGCTGATATATATAAAGCAAAGTCGCCTTCCGTGGAGCTCGGAGCTCTGAATGGAGCAA
CCGTTAAACACACGGTACAAATAAAGCGGTTTGATTTAAGGAGAACACATCCGGCGTCGACGGAAACACG
TGTGATTCGCGGGC
CCGTCGCCTTCATCGCTGTGTCTCCGGCGGGACATTTCTGCAGTTCAGCGAACACTGAGGAGGGAAACAC
TGAGGGAAATGCGCTGATATATATAAAGCAAAGTCGCCTTCCGTGGAGCTCGGAGCTCTGAATGGAGCAA
CCGTTAAACACACGGTACAAATAAAGCGGTTTGATTTAAGGAGAACACATCCGGCGTCGACGGAAACACG
TGTGATTCGCGGGC