ZFLNCG08306
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372793 | shield | normal | 30.53 |
| SRR658539 | bud | Gata5/6 morphant | 26.54 |
| SRR592701 | pineal gland | normal | 25.50 |
| SRR658533 | bud | normal | 25.05 |
| SRR658537 | bud | Gata6 morphant | 24.23 |
| ERR023146 | head kidney | normal | 23.49 |
| SRR1004788 | larvae | impdh1b morpholino | 19.33 |
| SRR658547 | 6 somite | Gata5/6 morphant | 18.75 |
| SRR748491 | germ ring | normal | 18.09 |
| SRR658541 | 6 somite | normal | 17.78 |
Express in tissues
Correlated coding gene
Download
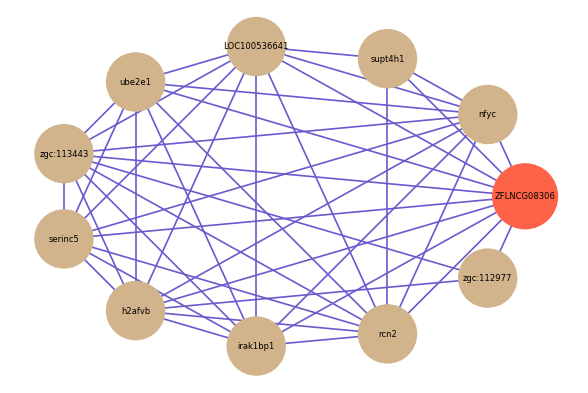
| gene | correlation coefficent |
|---|---|
| nfyc | 0.55 |
| supt4h1 | 0.54 |
| LOC100536641 | 0.54 |
| ube2e1 | 0.52 |
| zgc:113443 | 0.52 |
| zgc:112977 | 0.52 |
| serinc5 | 0.51 |
| h2afvb | 0.51 |
| irak1bp1 | 0.51 |
| rcn2 | 0.51 |
Gene Ontology
Download
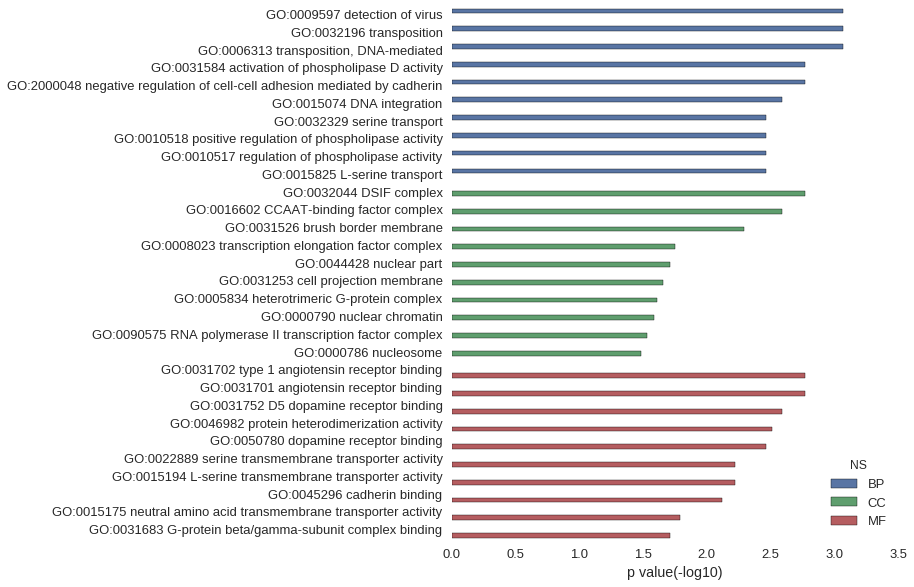
| GO | P value |
|---|---|
| GO:0009597 | 8.53e-04 |
| GO:0032196 | 8.53e-04 |
| GO:0006313 | 8.53e-04 |
| GO:0031584 | 1.70e-03 |
| GO:2000048 | 1.70e-03 |
| GO:0015074 | 2.56e-03 |
| GO:0032329 | 3.41e-03 |
| GO:0010518 | 3.41e-03 |
| GO:0010517 | 3.41e-03 |
| GO:0015825 | 3.41e-03 |
| GO:0032044 | 1.70e-03 |
| GO:0016602 | 2.56e-03 |
| GO:0031526 | 5.11e-03 |
| GO:0008023 | 1.78e-02 |
| GO:0044428 | 1.94e-02 |
| GO:0031253 | 2.20e-02 |
| GO:0005834 | 2.45e-02 |
| GO:0000790 | 2.61e-02 |
| GO:0090575 | 2.95e-02 |
| GO:0000786 | 3.28e-02 |
| GO:0031702 | 1.70e-03 |
| GO:0031701 | 1.70e-03 |
| GO:0031752 | 2.56e-03 |
| GO:0046982 | 3.09e-03 |
| GO:0050780 | 3.41e-03 |
| GO:0022889 | 5.96e-03 |
| GO:0015194 | 5.96e-03 |
| GO:0045296 | 7.65e-03 |
| GO:0015175 | 1.61e-02 |
| GO:0031683 | 1.94e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08306
TCTAATATACATCACGTCAACATGTACAACATGTAAATCAGTGGAGCTTGATGCGTCATCCACGTCTGGT
GTGAACGCAGCATTACACATTGCTCACTCACAAGGGAGAGGGTCCTCTGGCTTTTGCTGAAGATTTACTG
CTGATTACGTCACTGTTTAGCAGTAAATGTCAAGCAAAGGCCGGAGGACTCTCGCATTAATGCGAATTAT
ACACAATACCTAAATATGACCTGAAGAAACCCAGGCAATATCAG
TCTAATATACATCACGTCAACATGTACAACATGTAAATCAGTGGAGCTTGATGCGTCATCCACGTCTGGT
GTGAACGCAGCATTACACATTGCTCACTCACAAGGGAGAGGGTCCTCTGGCTTTTGCTGAAGATTTACTG
CTGATTACGTCACTGTTTAGCAGTAAATGTCAAGCAAAGGCCGGAGGACTCTCGCATTAATGCGAATTAT
ACACAATACCTAAATATGACCTGAAGAAACCCAGGCAATATCAG

