ZFLNCG08402
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023143 | swim bladder | normal | 1203.53 |
| ERR023145 | heart | normal | 543.09 |
| SRR592703 | pineal gland | normal | 354.90 |
| SRR592702 | pineal gland | normal | 290.01 |
| SRR1621206 | embryo | normal | 285.47 |
| SRR592701 | pineal gland | normal | 267.18 |
| SRR1049814 | 256 cell | normal | 213.67 |
| ERR023144 | brain | normal | 205.90 |
| SRR592700 | pineal gland | normal | 190.27 |
| ERR023146 | head kidney | normal | 141.73 |
Express in tissues
Correlated coding gene
Download
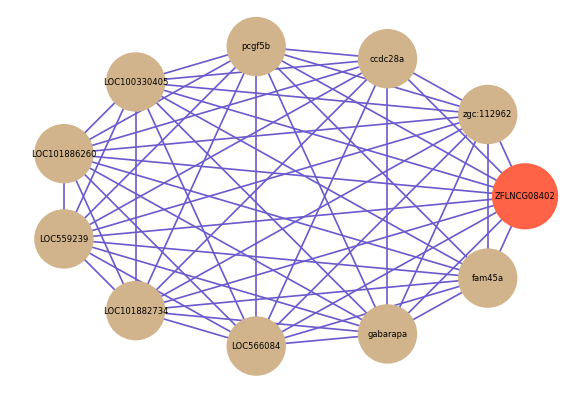
| gene | correlation coefficent |
|---|---|
| zgc:112962 | 0.64 |
| ccdc28a | 0.61 |
| pcgf5b | 0.61 |
| LOC100330405 | 0.61 |
| LOC101886260 | 0.61 |
| LOC559239 | 0.60 |
| LOC101882734 | 0.59 |
| LOC566084 | 0.59 |
| gabarapa | 0.58 |
| fam45a | 0.58 |
Gene Ontology
Download
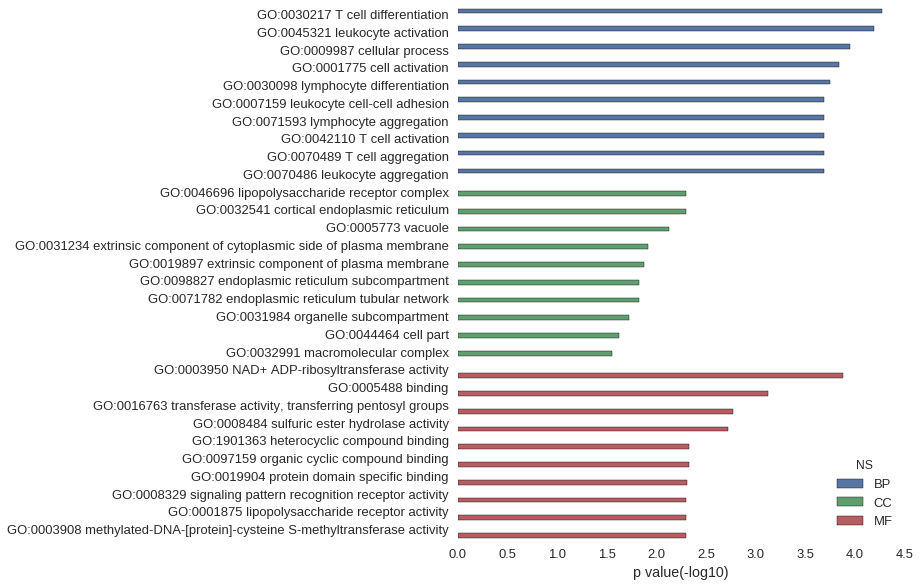
| GO | P value |
|---|---|
| GO:0030217 | 5.36e-05 |
| GO:0045321 | 6.33e-05 |
| GO:0009987 | 1.11e-04 |
| GO:0001775 | 1.45e-04 |
| GO:0030098 | 1.77e-04 |
| GO:0007159 | 2.03e-04 |
| GO:0071593 | 2.03e-04 |
| GO:0042110 | 2.03e-04 |
| GO:0070489 | 2.03e-04 |
| GO:0070486 | 2.03e-04 |
| GO:0046696 | 5.05e-03 |
| GO:0032541 | 5.05e-03 |
| GO:0005773 | 7.40e-03 |
| GO:0031234 | 1.20e-02 |
| GO:0019897 | 1.34e-02 |
| GO:0098827 | 1.51e-02 |
| GO:0071782 | 1.51e-02 |
| GO:0031984 | 1.90e-02 |
| GO:0044464 | 2.38e-02 |
| GO:0032991 | 2.80e-02 |
| GO:0003950 | 1.32e-04 |
| GO:0005488 | 7.48e-04 |
| GO:0016763 | 1.70e-03 |
| GO:0008484 | 1.89e-03 |
| GO:1901363 | 4.65e-03 |
| GO:0097159 | 4.69e-03 |
| GO:0019904 | 4.88e-03 |
| GO:0008329 | 5.05e-03 |
| GO:0001875 | 5.05e-03 |
| GO:0003908 | 5.05e-03 |
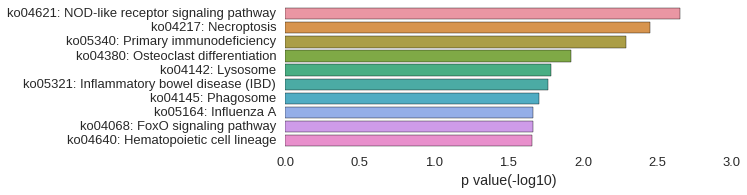
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08402
TTCGAGTCCCTCAATCATCCTCTGAGTGAAAATATGAATCTCACAATCATGCAGAAGATGTTGGAAAGGG
TTTAGATCTGCAAAAGATGCTGGAAAACTGAAGAATCTGCAGGAGATTGAGGATTTTTCTGTAGAACAGC
AGTCAGTTTAACTGTTCAGAACAAACAAGAGACTCATGAAGAACCATCTCAAGGCAGAAAAACACTCAGA
GATCATCAGTATACCACTAACGGTACTAAAAACCAGG
TTCGAGTCCCTCAATCATCCTCTGAGTGAAAATATGAATCTCACAATCATGCAGAAGATGTTGGAAAGGG
TTTAGATCTGCAAAAGATGCTGGAAAACTGAAGAATCTGCAGGAGATTGAGGATTTTTCTGTAGAACAGC
AGTCAGTTTAACTGTTCAGAACAAACAAGAGACTCATGAAGAACCATCTCAAGGCAGAAAAACACTCAGA
GATCATCAGTATACCACTAACGGTACTAAAAACCAGG