ZFLNCG08462
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1647683 | spleen | SVCV treatment | 42.96 |
| SRR1647684 | spleen | SVCV treatment | 37.71 |
| SRR1647682 | spleen | normal | 20.99 |
| SRR514030 | retina | id2a morpholino | 14.18 |
| SRR1205154 | 5dpf | transgenic sqET20 | 13.74 |
| SRR1205166 | 5dpf | transgenic sqET20 and neomycin treated 3h | 13.57 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 13.56 |
| SRR1104059 | heart | kctd10 mut | 12.76 |
| SRR535986 | larvae | nhsl1b mut | 11.90 |
| SRR594771 | endothelium | normal | 10.34 |
Express in tissues
Correlated coding gene
Download
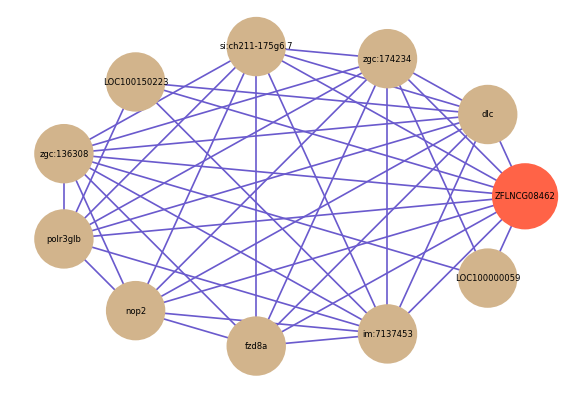
| gene | correlation coefficent |
|---|---|
| dlc | 0.74 |
| zgc:174234 | 0.59 |
| si:ch211-175g6.7 | 0.58 |
| LOC100150223 | 0.55 |
| zgc:136308 | 0.55 |
| polr3glb | 0.54 |
| nop2 | 0.54 |
| fzd8a | 0.54 |
| im:7137453 | 0.54 |
| LOC100000059 | 0.54 |
Gene Ontology
Download
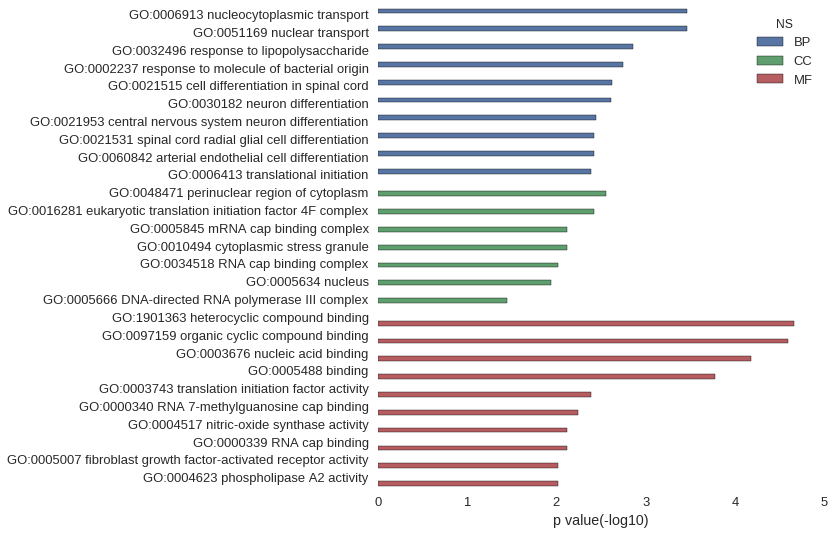
| GO | P value |
|---|---|
| GO:0006913 | 3.44e-04 |
| GO:0051169 | 3.44e-04 |
| GO:0032496 | 1.39e-03 |
| GO:0002237 | 1.80e-03 |
| GO:0021515 | 2.39e-03 |
| GO:0030182 | 2.46e-03 |
| GO:0021953 | 3.63e-03 |
| GO:0021531 | 3.83e-03 |
| GO:0060842 | 3.83e-03 |
| GO:0006413 | 4.10e-03 |
| GO:0048471 | 2.78e-03 |
| GO:0016281 | 3.83e-03 |
| GO:0005845 | 7.65e-03 |
| GO:0010494 | 7.65e-03 |
| GO:0034518 | 9.56e-03 |
| GO:0005634 | 1.16e-02 |
| GO:0005666 | 3.59e-02 |
| GO:1901363 | 2.20e-05 |
| GO:0097159 | 2.54e-05 |
| GO:0003676 | 6.55e-05 |
| GO:0005488 | 1.69e-04 |
| GO:0003743 | 4.10e-03 |
| GO:0000340 | 5.75e-03 |
| GO:0004517 | 7.65e-03 |
| GO:0000339 | 7.65e-03 |
| GO:0005007 | 9.56e-03 |
| GO:0004623 | 9.56e-03 |
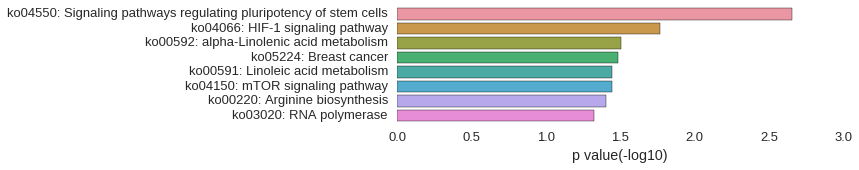
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08462
ATATGATGACAAATATATAAAAATTCTGTTTGGGTCATTAAAAAAGGGAAAGGAAATAGTGCAAATAATG
CAAGTTGTTCCACGGGTCTTCAAGTCTCTTGCTTGTCTTCACAAATTCATTATCCATATCAAGAAGTAGT
CCATTTAGTGAGCAGCATGAATAAATAAATAATATAATGCATGTGATATATACTCTCTTCACGTCTTCCT
TTTGTCCTTAGAGACTGTAAACATTTTTGTGGTCAGGCCCACTGGTGTTTGAGGTGCTCCAGATTGAAGA
ATTCTGCTATACCTGAGGAAGGACAGAAAGGAAAAAATGGCATTGATGCACGTTCTTTGAAAGCGTTCTG
CCTAAAAAAAATTAGCAAAAAAAAAAAAAAGACAGAAGATACTTACCTCAGTAGCAAACACACGTTGTTC
TATGTGTTCGGGAATGATGTACACAGGATGATACACTCCCTCTTTTGGATAGTTTAAAGGTGGAACCAGC
AATGTTGATTCAGAGTTTTTCCTGAAATTACGGAAACACATCCACGTTATGCATATAAAATGAAACAGAA
ATTGTGAACAGAAACTGAGGATTACTTGAACATGAACTTACTCTAGTTTGTTGTTTGTGTGCTTTTCATC
AATGCTCAGATTGTAGTCCACCATCTTCTGCTTATAGTTTGATTTGTCACTGGAATGTGTGTCTACAGAG
GTGGATCTGAGCGCCATATCTTTATTGGACACCTTAAATGGGCCACCAGGAATAAGGAAGGCCTCCTTCT
CTCTACCTAAAGTGGAGGTTGGGCTCAAAGAAATGCGATTATTGACAGAATCCAGGTTGTT
ATATGATGACAAATATATAAAAATTCTGTTTGGGTCATTAAAAAAGGGAAAGGAAATAGTGCAAATAATG
CAAGTTGTTCCACGGGTCTTCAAGTCTCTTGCTTGTCTTCACAAATTCATTATCCATATCAAGAAGTAGT
CCATTTAGTGAGCAGCATGAATAAATAAATAATATAATGCATGTGATATATACTCTCTTCACGTCTTCCT
TTTGTCCTTAGAGACTGTAAACATTTTTGTGGTCAGGCCCACTGGTGTTTGAGGTGCTCCAGATTGAAGA
ATTCTGCTATACCTGAGGAAGGACAGAAAGGAAAAAATGGCATTGATGCACGTTCTTTGAAAGCGTTCTG
CCTAAAAAAAATTAGCAAAAAAAAAAAAAAGACAGAAGATACTTACCTCAGTAGCAAACACACGTTGTTC
TATGTGTTCGGGAATGATGTACACAGGATGATACACTCCCTCTTTTGGATAGTTTAAAGGTGGAACCAGC
AATGTTGATTCAGAGTTTTTCCTGAAATTACGGAAACACATCCACGTTATGCATATAAAATGAAACAGAA
ATTGTGAACAGAAACTGAGGATTACTTGAACATGAACTTACTCTAGTTTGTTGTTTGTGTGCTTTTCATC
AATGCTCAGATTGTAGTCCACCATCTTCTGCTTATAGTTTGATTTGTCACTGGAATGTGTGTCTACAGAG
GTGGATCTGAGCGCCATATCTTTATTGGACACCTTAAATGGGCCACCAGGAATAAGGAAGGCCTCCTTCT
CTCTACCTAAAGTGGAGGTTGGGCTCAAAGAAATGCGATTATTGACAGAATCCAGGTTGTT