ZFLNCG08475
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562530 | liver | normal | 1.97 |
| SRR1035239 | liver | transgenic mCherry control | 1.03 |
| SRR527834 | head | normal | 0.73 |
| ERR023146 | head kidney | normal | 0.71 |
| SRR633544 | larvae | exposed to cold stress | 0.61 |
| SRR1167757 | embryo | mpeg1 morpholino and bacterial infection | 0.39 |
| SRR1005529 | embryo | control morpholino | 0.36 |
| SRR801554 | embryo | control morpholino | 0.35 |
| SRR527833 | 5 dpf | normal | 0.33 |
| SRR1039573 | gastrointestinal | diet 0.75 NPM | 0.32 |
Express in tissues
Correlated coding gene
Download
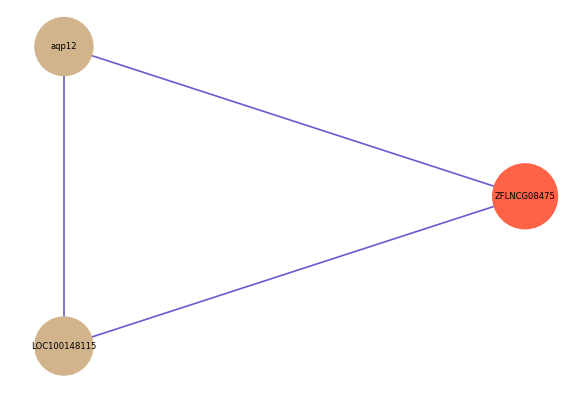
| gene | correlation coefficent |
|---|---|
| aqp12 | 0.50 |
| LOC100148115 | 0.50 |
Gene Ontology
Download
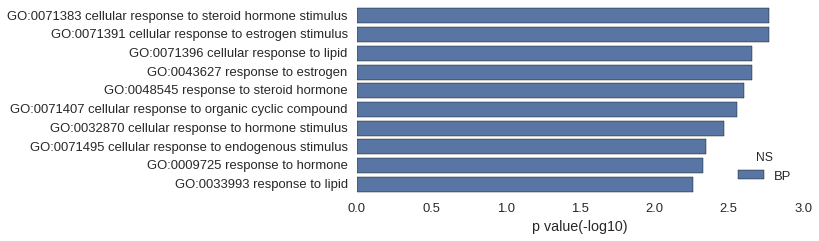
| GO | P value |
|---|---|
| GO:0071383 | 1.71e-03 |
| GO:0071391 | 1.71e-03 |
| GO:0071396 | 2.20e-03 |
| GO:0043627 | 2.20e-03 |
| GO:0048545 | 2.49e-03 |
| GO:0071407 | 2.77e-03 |
| GO:0032870 | 3.41e-03 |
| GO:0071495 | 4.48e-03 |
| GO:0009725 | 4.69e-03 |
| GO:0033993 | 5.54e-03 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08475
CCTTATTACCAAAACATTTTTCAATGTCTAAACTTTTGCTAGCGGCAATAATATTTTCACTGGGCAGAAC
ACAGTACGCTAGAATCCGCACTACAGGAGCCACATCTGCACCAACAGACAGTTCGAAGGACATTGTGCCA
CTTGCTACTCCATTTGAAGACTTCACTTCAACTTTCTCATACCCATGATGAACAATCACTCCTTTGGACA
AGACACTGTTATAAACTGACCAGTGCAGTAAATTAATGCTGCAAACAAAATAAATAAATTAGCCATCGCT
TTAAGAATGTGCACAAGTACTCACTATATAGACAATGTCAGTATTGAAGTCTTCAACCGTCTCTCCAACA
AAATAATACTTTACAGTGGCTGTAATCTCAGTGCCACACTTTATCGGCTGCTCAATATTCTCTATTATCA
GTTTACTAAATGTTGGGGAGTATGGAGTTGCGGTCTGGAAGAGTTTAACTGTTTTTTTATCTCTAGAGAA
GTAAGGTGTTTTGTACTGACGAAAACGGGTATCTGGATACACATTTGC
CCTTATTACCAAAACATTTTTCAATGTCTAAACTTTTGCTAGCGGCAATAATATTTTCACTGGGCAGAAC
ACAGTACGCTAGAATCCGCACTACAGGAGCCACATCTGCACCAACAGACAGTTCGAAGGACATTGTGCCA
CTTGCTACTCCATTTGAAGACTTCACTTCAACTTTCTCATACCCATGATGAACAATCACTCCTTTGGACA
AGACACTGTTATAAACTGACCAGTGCAGTAAATTAATGCTGCAAACAAAATAAATAAATTAGCCATCGCT
TTAAGAATGTGCACAAGTACTCACTATATAGACAATGTCAGTATTGAAGTCTTCAACCGTCTCTCCAACA
AAATAATACTTTACAGTGGCTGTAATCTCAGTGCCACACTTTATCGGCTGCTCAATATTCTCTATTATCA
GTTTACTAAATGTTGGGGAGTATGGAGTTGCGGTCTGGAAGAGTTTAACTGTTTTTTTATCTCTAGAGAA
GTAAGGTGTTTTGTACTGACGAAAACGGGTATCTGGATACACATTTGC
