ZFLNCG08702
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 136.37 |
| SRR372802 | 5 dpf | normal | 105.89 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 104.76 |
| SRR065196 | 3 dpf | normal | 92.50 |
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 90.45 |
| SRR065197 | 3 dpf | U1C knockout | 86.62 |
| SRR1205163 | 5dpf | transgenic sqET20 and neomycin treated 3h and GFP+ | 81.27 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 80.65 |
| SRR726542 | 5 dpf | infection with control | 74.50 |
| SRR519732 | 7 dpf | FETOH treatment | 63.16 |
Express in tissues
Correlated coding gene
Download
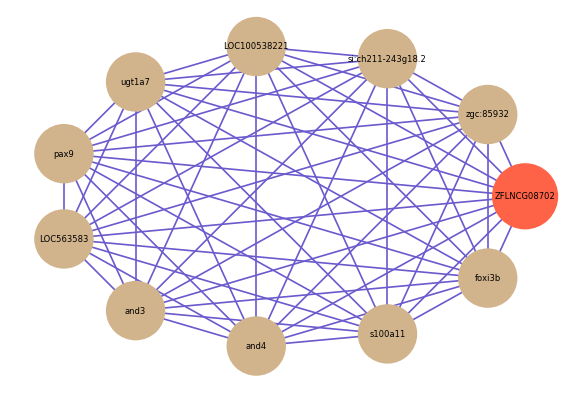
| gene | correlation coefficent |
|---|---|
| zgc:85932 | 0.88 |
| si:ch211-243g18.2 | 0.76 |
| LOC100538221 | 0.76 |
| ugt1a7 | 0.76 |
| pax9 | 0.76 |
| LOC563583 | 0.75 |
| and3 | 0.74 |
| and4 | 0.74 |
| s100a11 | 0.74 |
| foxi3b | 0.74 |
Gene Ontology
Download
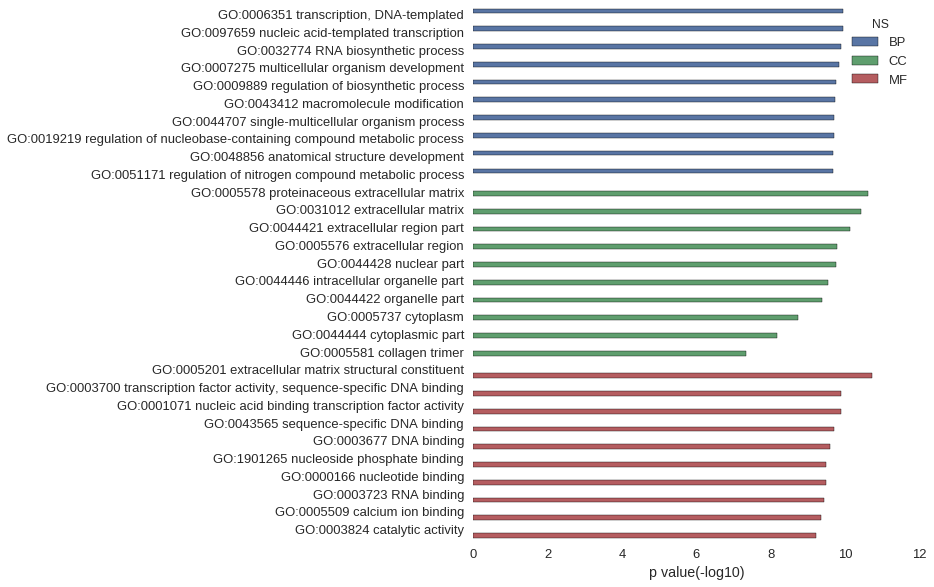
| GO | P value |
|---|---|
| GO:0006351 | 1.18e-10 |
| GO:0097659 | 1.18e-10 |
| GO:0032774 | 1.33e-10 |
| GO:0007275 | 1.50e-10 |
| GO:0009889 | 1.75e-10 |
| GO:0043412 | 1.86e-10 |
| GO:0044707 | 1.97e-10 |
| GO:0019219 | 1.97e-10 |
| GO:0048856 | 2.10e-10 |
| GO:0051171 | 2.19e-10 |
| GO:0005578 | 2.37e-11 |
| GO:0031012 | 3.85e-11 |
| GO:0044421 | 7.50e-11 |
| GO:0005576 | 1.67e-10 |
| GO:0044428 | 1.76e-10 |
| GO:0044446 | 2.96e-10 |
| GO:0044422 | 4.09e-10 |
| GO:0005737 | 1.80e-09 |
| GO:0044444 | 6.71e-09 |
| GO:0005581 | 4.51e-08 |
| GO:0005201 | 1.93e-11 |
| GO:0003700 | 1.32e-10 |
| GO:0001071 | 1.32e-10 |
| GO:0043565 | 2.02e-10 |
| GO:0003677 | 2.59e-10 |
| GO:1901265 | 3.22e-10 |
| GO:0000166 | 3.22e-10 |
| GO:0003723 | 3.80e-10 |
| GO:0005509 | 4.59e-10 |
| GO:0003824 | 6.14e-10 |
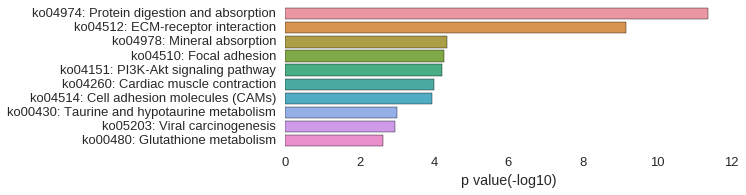
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08702
GTTATTGCGTGAGTCAGAGATGAACTCATTATCTTGTGATGATTGTGATTCATTTACTGTTGAGCTCATG
AATATGCATGAGTGTTTGTAACTCTTCTCTTGATTTGTAGTTCCTGCACATGACGTTGTACAACTGATCC
CGGAGCAGCGAGAAGCTCCACATCTGCTGTAACTGTAACTGCTGTATGTGTGTGTGCGTGTGGTCAGTGC
TGTAACACACGGACATGGCGAGGAGCAGCTACTGTAAATAACACACTGCACTGGGGAACACTGCAGGTTT
AGTGTACTGCAGACAGTGGACGGGTGATTGCTGAATAAACAGATATTTTAAAGAACATCAGCAAAATTGA
TGTAATATTT
GTTATTGCGTGAGTCAGAGATGAACTCATTATCTTGTGATGATTGTGATTCATTTACTGTTGAGCTCATG
AATATGCATGAGTGTTTGTAACTCTTCTCTTGATTTGTAGTTCCTGCACATGACGTTGTACAACTGATCC
CGGAGCAGCGAGAAGCTCCACATCTGCTGTAACTGTAACTGCTGTATGTGTGTGTGCGTGTGGTCAGTGC
TGTAACACACGGACATGGCGAGGAGCAGCTACTGTAAATAACACACTGCACTGGGGAACACTGCAGGTTT
AGTGTACTGCAGACAGTGGACGGGTGATTGCTGAATAAACAGATATTTTAAAGAACATCAGCAAAATTGA
TGTAATATTT