ZFLNCG08712
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 58.94 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 51.83 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 44.02 |
| SRR516123 | skin | male and 3.5 year | 42.73 |
| SRR527835 | tail | normal | 29.15 |
| SRR516125 | skin | male and 3.5 year | 26.11 |
| SRR516121 | skin | male and 3.5 year | 23.13 |
| SRR1299128 | caudal fin | Three days time after treatment | 19.49 |
| SRR372787 | 2-4 cell | normal | 19.35 |
| SRR1299127 | caudal fin | Two days time after treatment | 19.00 |
Express in tissues
Correlated coding gene
Download
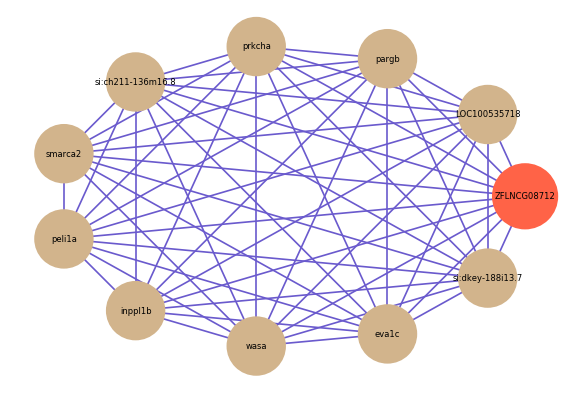
| gene | correlation coefficent |
|---|---|
| LOC100535718 | 0.84 |
| pargb | 0.74 |
| prkcha | 0.66 |
| si:ch211-136m16.8 | 0.64 |
| smarca2 | 0.62 |
| peli1a | 0.61 |
| inppl1b | 0.61 |
| wasa | 0.61 |
| eva1c | 0.61 |
| si:dkey-188i13.7 | 0.61 |
Gene Ontology
Download
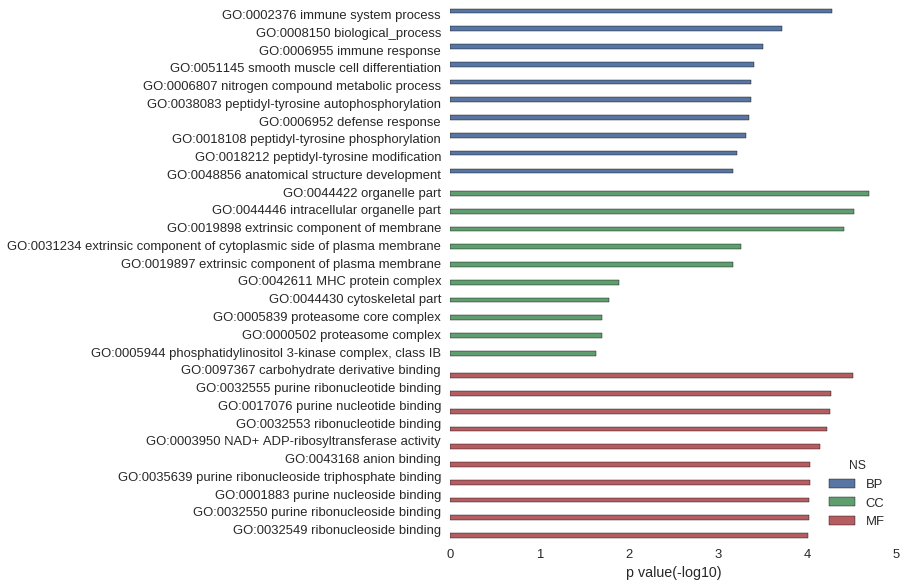
| GO | P value |
|---|---|
| GO:0002376 | 5.23e-05 |
| GO:0008150 | 1.91e-04 |
| GO:0006955 | 3.16e-04 |
| GO:0051145 | 3.97e-04 |
| GO:0006807 | 4.22e-04 |
| GO:0038083 | 4.28e-04 |
| GO:0006952 | 4.45e-04 |
| GO:0018108 | 4.84e-04 |
| GO:0018212 | 6.14e-04 |
| GO:0048856 | 6.71e-04 |
| GO:0044422 | 2.05e-05 |
| GO:0044446 | 2.97e-05 |
| GO:0019898 | 3.84e-05 |
| GO:0031234 | 5.46e-04 |
| GO:0019897 | 6.87e-04 |
| GO:0042611 | 1.27e-02 |
| GO:0044430 | 1.66e-02 |
| GO:0005839 | 2.00e-02 |
| GO:0000502 | 2.01e-02 |
| GO:0005944 | 2.30e-02 |
| GO:0097367 | 3.10e-05 |
| GO:0032555 | 5.45e-05 |
| GO:0017076 | 5.54e-05 |
| GO:0032553 | 6.04e-05 |
| GO:0003950 | 7.27e-05 |
| GO:0043168 | 9.24e-05 |
| GO:0035639 | 9.37e-05 |
| GO:0001883 | 9.46e-05 |
| GO:0032550 | 9.46e-05 |
| GO:0032549 | 9.73e-05 |
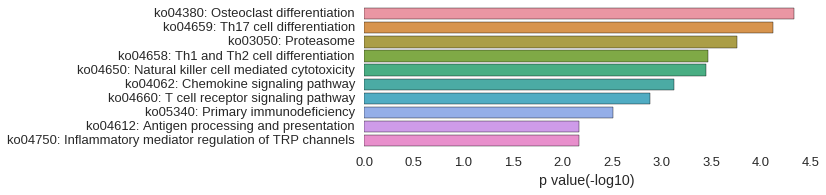
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08712
TGATAGCCACGATTAACACACACATCCAGACCACCAGTTAACACACACACAGATTCTAGATTGCCACAAT
GAACACACACACACAGAGTGATTTTATATAGCCACGATAAGCACATACACACACAGTGATTCTTGATAGC
CACCATAAACACACACAGAAGGATTCTAGATAGCCATAACACACACACACACACAAGTCCAGACCACCAC
GATGAAAACACACATACACAGAGTGATGAACGCACAAAGAAAACACACACACATACATTAACACATGTCT
CCCCTCGACCACACATACTGTACATACATTCAGATTTTCACCTAAACACTCTTAATAATGGACACACACA
CACACACACACACACACCTCGGTTGTGTGTTTGTGTGCAGTGTTTGGATTGTCTGCCATATAGCTTTCTC
TGTCTCTGTCGTCTTCTCCTCATTCAGTGATTGATCAGTAAAGTCACATGAGCTCACCTAATGTACATGA
ATATTAATGAAGCGCTTGCTCTCTGAGACCAATCGGATGCCAGAGTGGGCGGTTCCTCCGCTCCAGTCCA
GTGTTATTTATTAAATGAACTGTCTTTGTGTTTTTATACGTGTGCTGGAGCTGCTGTATGCTTTCTCTTT
CTGCTCTGTTCAAGTGAGTAGCTTTGAACACACACACACACACACACACACACACACACACATGCAAAGC
ACATCTGTTTACATTTTATAGCAAAGT
TGATAGCCACGATTAACACACACATCCAGACCACCAGTTAACACACACACAGATTCTAGATTGCCACAAT
GAACACACACACACAGAGTGATTTTATATAGCCACGATAAGCACATACACACACAGTGATTCTTGATAGC
CACCATAAACACACACAGAAGGATTCTAGATAGCCATAACACACACACACACACAAGTCCAGACCACCAC
GATGAAAACACACATACACAGAGTGATGAACGCACAAAGAAAACACACACACATACATTAACACATGTCT
CCCCTCGACCACACATACTGTACATACATTCAGATTTTCACCTAAACACTCTTAATAATGGACACACACA
CACACACACACACACACCTCGGTTGTGTGTTTGTGTGCAGTGTTTGGATTGTCTGCCATATAGCTTTCTC
TGTCTCTGTCGTCTTCTCCTCATTCAGTGATTGATCAGTAAAGTCACATGAGCTCACCTAATGTACATGA
ATATTAATGAAGCGCTTGCTCTCTGAGACCAATCGGATGCCAGAGTGGGCGGTTCCTCCGCTCCAGTCCA
GTGTTATTTATTAAATGAACTGTCTTTGTGTTTTTATACGTGTGCTGGAGCTGCTGTATGCTTTCTCTTT
CTGCTCTGTTCAAGTGAGTAGCTTTGAACACACACACACACACACACACACACACACACACATGCAAAGC
ACATCTGTTTACATTTTATAGCAAAGT