ZFLNCG08743
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR886457 | 512 cell | 4-thio-UTP metabolic labeling | 583.08 |
| SRR886455 | 128 cell | 4-thio-UTP metabolic labeling | 489.61 |
| SRR886456 | 256 cell | 4-thio-UTP metabolic labeling | 338.40 |
| SRR1035978 | 13 hpf | rx3-/- | 328.44 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 300.89 |
| SRR658533 | bud | normal | 255.95 |
| SRR1035984 | 13 hpf | normal | 238.74 |
| SRR658539 | bud | Gata5/6 morphant | 234.58 |
| SRR658545 | 6 somite | Gata6 morphant | 223.68 |
| SRR658537 | bud | Gata6 morphant | 223.36 |
Express in tissues
Correlated coding gene
Download
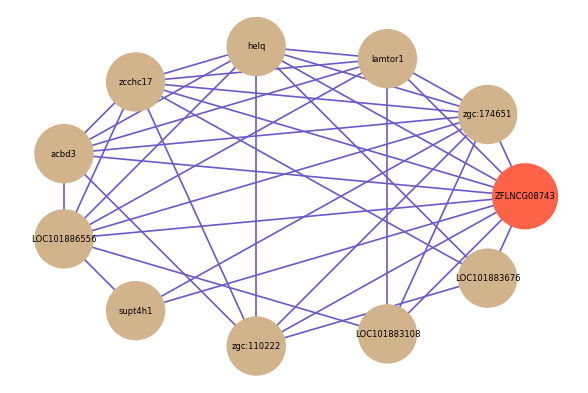
| gene | correlation coefficent |
|---|---|
| zgc:174651 | 0.66 |
| lamtor1 | 0.63 |
| helq | 0.60 |
| zcchc17 | 0.58 |
| acbd3 | 0.55 |
| LOC101886556 | 0.54 |
| supt4h1 | 0.54 |
| zgc:110222 | 0.54 |
| LOC101883108 | 0.53 |
| LOC101883676 | 0.52 |
Gene Ontology
Download
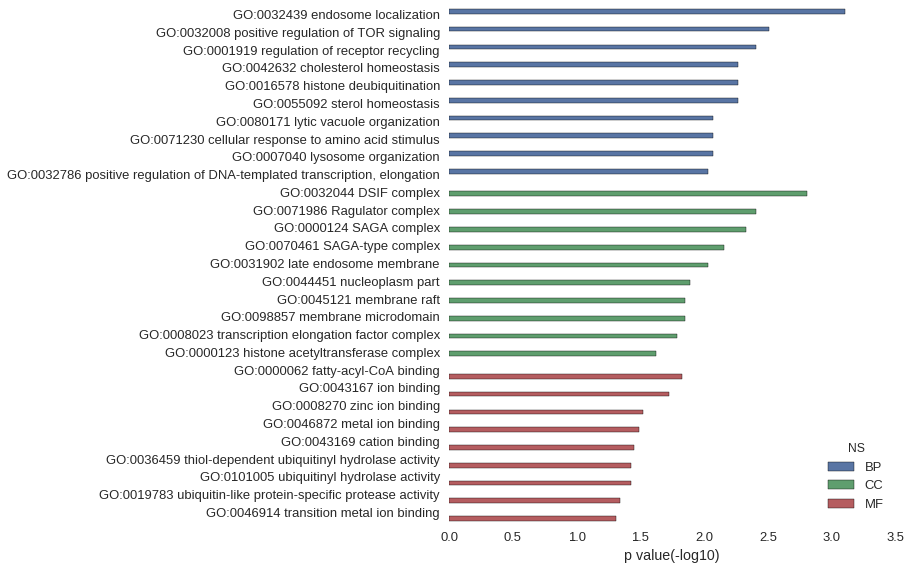
| GO | P value |
|---|---|
| GO:0032439 | 7.82e-04 |
| GO:0032008 | 3.12e-03 |
| GO:0001919 | 3.90e-03 |
| GO:0042632 | 5.46e-03 |
| GO:0016578 | 5.46e-03 |
| GO:0055092 | 5.46e-03 |
| GO:0080171 | 8.57e-03 |
| GO:0071230 | 8.57e-03 |
| GO:0007040 | 8.57e-03 |
| GO:0032786 | 9.34e-03 |
| GO:0032044 | 1.56e-03 |
| GO:0071986 | 3.90e-03 |
| GO:0000124 | 4.68e-03 |
| GO:0070461 | 7.02e-03 |
| GO:0031902 | 9.34e-03 |
| GO:0044451 | 1.28e-02 |
| GO:0045121 | 1.40e-02 |
| GO:0098857 | 1.40e-02 |
| GO:0008023 | 1.63e-02 |
| GO:0000123 | 2.40e-02 |
| GO:0000062 | 1.48e-02 |
| GO:0043167 | 1.90e-02 |
| GO:0008270 | 3.04e-02 |
| GO:0046872 | 3.27e-02 |
| GO:0043169 | 3.54e-02 |
| GO:0036459 | 3.77e-02 |
| GO:0101005 | 3.77e-02 |
| GO:0019783 | 4.59e-02 |
| GO:0046914 | 4.95e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08743
GCTTGTCATCTTCATCCTCATCATCATCACTGCCTAAAGCCTTTCGTTTGGGCCCTTCATCATCTTCATC
ATCACTGGCTAGAGCTCTCCGTTTGGGTCTTTCATCTTCATCATCACTATCTAAAGCCTTTCGTTTGGGC
CCTTCATCTTCATCATCACTAGCTAGAACTTTTCGTTTGGGTCTTTCATCTTCATCATCATCACTATCTA
AAGCTTTTGATTTGGGCCCTCCATCATCATCATCACTACCTAAAGCCTTTCGTTTAGGCCCTTCATCTTC
ATCATCATCACTACCTAAAGCCTTTCGTTTGGGCCTTTTATCATCATCATCGTCATCATCATCACTATCT
AAAGCCTTTCGTTTAGGACGTTTATCATCTTCATCGCTGTTTAAAGCCTTTTGTTTAGGCTCTTCATCCT
CATCATCATCACTACCTAAAGCCTTTCGTTTGGGCCTTTCATCTTCATCATCACTAGCCAGAGCCTTTCT
TTGAGGCCTTCCATCATCTTCATCATCACTACCTAGAGCCCTTCTTTTAGGCCTTT
GCTTGTCATCTTCATCCTCATCATCATCACTGCCTAAAGCCTTTCGTTTGGGCCCTTCATCATCTTCATC
ATCACTGGCTAGAGCTCTCCGTTTGGGTCTTTCATCTTCATCATCACTATCTAAAGCCTTTCGTTTGGGC
CCTTCATCTTCATCATCACTAGCTAGAACTTTTCGTTTGGGTCTTTCATCTTCATCATCATCACTATCTA
AAGCTTTTGATTTGGGCCCTCCATCATCATCATCACTACCTAAAGCCTTTCGTTTAGGCCCTTCATCTTC
ATCATCATCACTACCTAAAGCCTTTCGTTTGGGCCTTTTATCATCATCATCGTCATCATCATCACTATCT
AAAGCCTTTCGTTTAGGACGTTTATCATCTTCATCGCTGTTTAAAGCCTTTTGTTTAGGCTCTTCATCCT
CATCATCATCACTACCTAAAGCCTTTCGTTTGGGCCTTTCATCTTCATCATCACTAGCCAGAGCCTTTCT
TTGAGGCCTTCCATCATCTTCATCATCACTACCTAGAGCCCTTCTTTTAGGCCTTT