ZFLNCG08754
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 12.78 |
| SRR592698 | pineal gland | normal | 11.57 |
| SRR527834 | head | normal | 11.53 |
| SRR592702 | pineal gland | normal | 9.84 |
| ERR594417 | retina | normal | 8.84 |
| SRR748488 | sperm | normal | 8.80 |
| SRR592701 | pineal gland | normal | 8.56 |
| SRR1648854 | brain | normal | 8.47 |
| SRR592699 | pineal gland | normal | 8.29 |
| SRR516135 | skin | male and 5 month | 7.70 |
Express in tissues
Correlated coding gene
Download
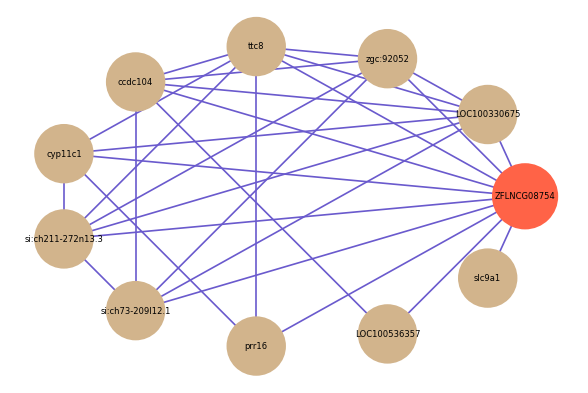
| gene | correlation coefficent |
|---|---|
| LOC100330675 | 0.77 |
| zgc:92052 | 0.63 |
| prr16 | 0.58 |
| ttc8 | 0.58 |
| ccdc104 | 0.58 |
| slc9a1 | 0.57 |
| cyp11c1 | 0.57 |
| LOC100536357 | 0.57 |
| si:ch211-272n13.3 | 0.56 |
| si:ch73-209l12.1 | 0.55 |
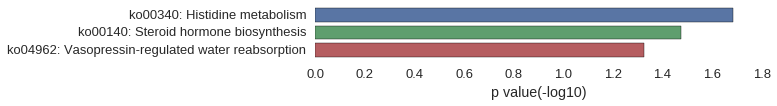
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0008212 | 5.69e-04 |
| GO:0006705 | 5.69e-04 |
| GO:0032341 | 5.69e-04 |
| GO:0032342 | 5.69e-04 |
| GO:0042384 | 1.08e-03 |
| GO:0006700 | 1.14e-03 |
| GO:0006704 | 1.14e-03 |
| GO:0044782 | 1.36e-03 |
| GO:0030031 | 1.47e-03 |
| GO:0034650 | 1.70e-03 |
| GO:0030991 | 2.27e-03 |
| GO:0005869 | 3.41e-03 |
| GO:0000777 | 4.54e-03 |
| GO:0005856 | 9.65e-03 |
| GO:0000776 | 1.30e-02 |
| GO:0000775 | 1.36e-02 |
| GO:0098687 | 2.20e-02 |
| GO:0005929 | 4.40e-02 |
| GO:0005875 | 4.73e-02 |
| GO:0004507 | 5.69e-04 |
| GO:0047783 | 5.69e-04 |
| GO:0016713 | 1.14e-03 |
| GO:0005451 | 5.11e-03 |
| GO:0015385 | 5.11e-03 |
| GO:0015299 | 6.24e-03 |
| GO:0016709 | 9.06e-03 |
| GO:0016831 | 1.30e-02 |
| GO:0015491 | 1.36e-02 |
| GO:0015298 | 1.47e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08754
TATGATCTTTCTGTGTGTGTGTGTGTTTGTCACCTTAGTGGTGTGTATCCTGAGAAGGTTGTGTGTGTTC
TGCTGGTTTCTGTGGAAAAAAAATGATGTTCAAATGACACGATCACTCAGATGATGTCAGGTGTGTGTGT
GTGTGTGTGTGTGTGTGCGTCACCTGTGCTCAGGCCGCTCCAACGGTTCTGCATTGATTCTGTGGGTCTG
TGAGCGAGTCCATTTGACAGGGCTGAGGAGGGGTGTGTAGGCCATGCCCACGAGGGGCGGGGCAGCAGGG
GTGTGGTCAGAGGAGGCGGGGCTTGTTTGTGTGTTACTGAACTCAGAGCTCCTAACACAGAGTTGAGCTG
ACCTGAGATCAGCTGCAGAGAGTCGGCCAACTGCTGCACCTTCACTGGAACTGAGGAGACGCAGAGGTCA
GTGTGTGTACGTGTGTGTGTGTGTGTGTGTGTGTCTTACCTGTTCCCTCTGGGCTGTAAACACTGCTCAT
CTCTGATTCGGTCACATCAAACGTCACTTTACGATCAGCTGACCTCTCGCCGTCATCACATGACACCTGA
TATACACACACACAC
TATGATCTTTCTGTGTGTGTGTGTGTTTGTCACCTTAGTGGTGTGTATCCTGAGAAGGTTGTGTGTGTTC
TGCTGGTTTCTGTGGAAAAAAAATGATGTTCAAATGACACGATCACTCAGATGATGTCAGGTGTGTGTGT
GTGTGTGTGTGTGTGTGCGTCACCTGTGCTCAGGCCGCTCCAACGGTTCTGCATTGATTCTGTGGGTCTG
TGAGCGAGTCCATTTGACAGGGCTGAGGAGGGGTGTGTAGGCCATGCCCACGAGGGGCGGGGCAGCAGGG
GTGTGGTCAGAGGAGGCGGGGCTTGTTTGTGTGTTACTGAACTCAGAGCTCCTAACACAGAGTTGAGCTG
ACCTGAGATCAGCTGCAGAGAGTCGGCCAACTGCTGCACCTTCACTGGAACTGAGGAGACGCAGAGGTCA
GTGTGTGTACGTGTGTGTGTGTGTGTGTGTGTGTCTTACCTGTTCCCTCTGGGCTGTAAACACTGCTCAT
CTCTGATTCGGTCACATCAAACGTCACTTTACGATCAGCTGACCTCTCGCCGTCATCACATGACACCTGA
TATACACACACACAC