ZFLNCG08862
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514028 | retina | control morpholino | 2.46 |
| SRR797911 | embryo | prdm1-/- | 1.73 |
| SRR535986 | larvae | nhsl1b mut | 1.40 |
| ERR594418 | retina | transgenic flk1 and develop tumors | 1.23 |
| SRR1648854 | brain | normal | 0.87 |
| SRR797917 | embryo | pbx2/4-MO and prdm1-/- | 0.71 |
| SRR519737 | 2 dpf | VD3 treatment | 0.68 |
| SRR038627 | embryo | control morpholino | 0.68 |
| SRR065197 | 3 dpf | U1C knockout | 0.61 |
| SRR610741 | 3 dpf | express Ras | 0.53 |
Express in tissues
Correlated coding gene
Download
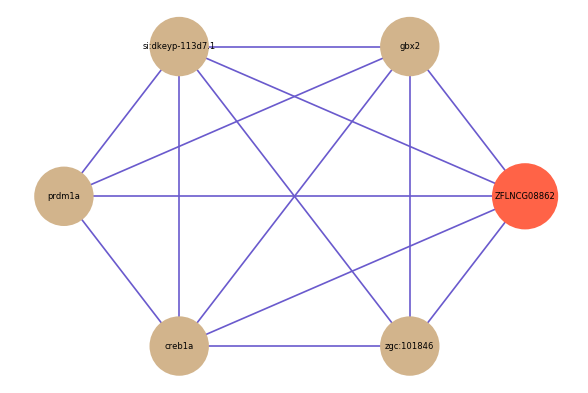
| gene | correlation coefficent |
|---|---|
| gbx2 | 0.52 |
| si:dkeyp-113d7.1 | 0.52 |
| prdm1a | 0.51 |
| creb1a | 0.50 |
| zgc:101846 | 0.50 |
Gene Ontology
Download
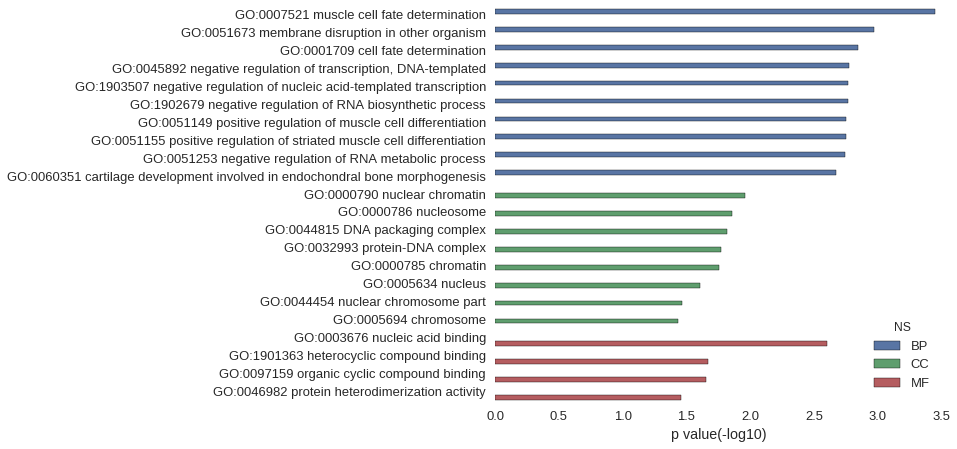
| GO | P value |
|---|---|
| GO:0007521 | 3.55e-04 |
| GO:0051673 | 1.07e-03 |
| GO:0001709 | 1.42e-03 |
| GO:0045892 | 1.69e-03 |
| GO:1903507 | 1.71e-03 |
| GO:1902679 | 1.71e-03 |
| GO:0051149 | 1.78e-03 |
| GO:0051155 | 1.78e-03 |
| GO:0051253 | 1.82e-03 |
| GO:0060351 | 2.13e-03 |
| GO:0000790 | 1.10e-02 |
| GO:0000786 | 1.38e-02 |
| GO:0044815 | 1.52e-02 |
| GO:0032993 | 1.69e-02 |
| GO:0000785 | 1.76e-02 |
| GO:0005634 | 2.47e-02 |
| GO:0044454 | 3.43e-02 |
| GO:0005694 | 3.68e-02 |
| GO:0003676 | 2.50e-03 |
| GO:1901363 | 2.16e-02 |
| GO:0097159 | 2.24e-02 |
| GO:0046982 | 3.47e-02 |
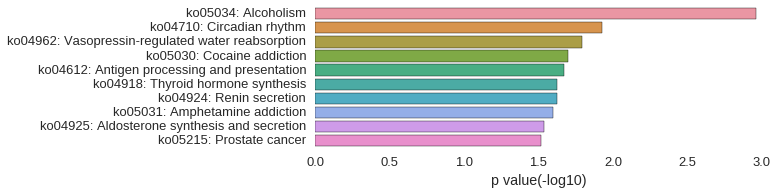
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08862
AAACACTGGAGCGGCAGGAGAGAGAGAAGCAGACCTGAGGATAAAGTGGGGCGTGTGACGGATGGCCCAA
GTACTAAAGGAGGAGACGTGGACGCTGAGAGCCAGGGGACAGGGGCAGGTCGAGAGGCTGCTAATCAGAG
GGACCCCGCAGAGCACACAAGACCCTAGGCCTGACGCAGGGGCACGATGGCCCCCAGCCTGCTCCTGCCG
TGCAGTGTAGACAGTCCCGCTGAGAAAGCAGCGCGCACAAAAGAGCTGCGGCCCGGCAGGGAGCGCAGCG
CTGAGCCGCAGGTTAGCATGACAAAGGCCCTGTAATGGAGTCTACCACACCCCAGCCCTCAGCTAAACAA
ACAGCCACCCTCAGATTCCATTAAGATAAGAGCAGAACACACACTCATCTCTGCATGACGACTAACACAC
ACTTAGCTACTGCACAAACACACACATTGGGAGGAAGACAAATGAAGAAAGTTGCACTCAAATCCATGCT
TTTTTTTGTCCACAGATCATCATGAAGAATGGAGACACAATGGAGACAGTTTGACTTTGTTTTTCAAAGT
TTGTTCAAGTTGAGACCAAGAACGTTCTCTTTGATTTAGACAAGATGCTTCTAAAAATACTATTTTTGAA
ACGAAAATGCTACAAAACAGACAAACT
AAACACTGGAGCGGCAGGAGAGAGAGAAGCAGACCTGAGGATAAAGTGGGGCGTGTGACGGATGGCCCAA
GTACTAAAGGAGGAGACGTGGACGCTGAGAGCCAGGGGACAGGGGCAGGTCGAGAGGCTGCTAATCAGAG
GGACCCCGCAGAGCACACAAGACCCTAGGCCTGACGCAGGGGCACGATGGCCCCCAGCCTGCTCCTGCCG
TGCAGTGTAGACAGTCCCGCTGAGAAAGCAGCGCGCACAAAAGAGCTGCGGCCCGGCAGGGAGCGCAGCG
CTGAGCCGCAGGTTAGCATGACAAAGGCCCTGTAATGGAGTCTACCACACCCCAGCCCTCAGCTAAACAA
ACAGCCACCCTCAGATTCCATTAAGATAAGAGCAGAACACACACTCATCTCTGCATGACGACTAACACAC
ACTTAGCTACTGCACAAACACACACATTGGGAGGAAGACAAATGAAGAAAGTTGCACTCAAATCCATGCT
TTTTTTTGTCCACAGATCATCATGAAGAATGGAGACACAATGGAGACAGTTTGACTTTGTTTTTCAAAGT
TTGTTCAAGTTGAGACCAAGAACGTTCTCTTTGATTTAGACAAGATGCTTCTAAAAATACTATTTTTGAA
ACGAAAATGCTACAAAACAGACAAACT