ZFLNCG08909
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR594418 | retina | transgenic flk1 and develop tumors | 44.82 |
| ERR594417 | retina | normal | 35.51 |
| ERR594416 | retina | transgenic flk1 | 22.52 |
| SRR658537 | bud | Gata6 morphant | 18.26 |
| SRR514030 | retina | id2a morpholino | 18.16 |
| ERR023144 | brain | normal | 17.28 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 16.69 |
| SRR514028 | retina | control morpholino | 15.07 |
| SRR1205154 | 5dpf | transgenic sqET20 | 13.57 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 13.56 |
Express in tissues
Correlated coding gene
Download
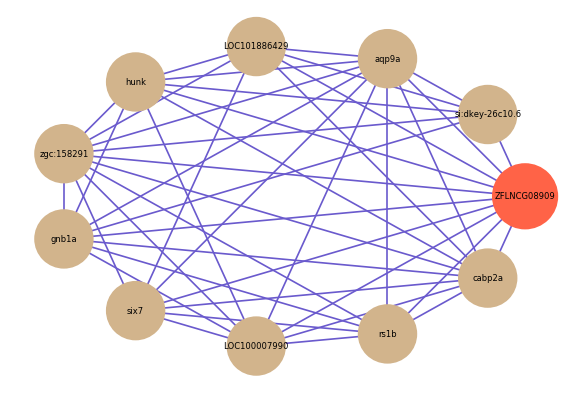
| gene | correlation coefficent |
|---|---|
| si:dkey-26c10.6 | 0.67 |
| aqp9a | 0.62 |
| LOC101886429 | 0.58 |
| hunk | 0.56 |
| six7 | 0.56 |
| LOC100007990 | 0.56 |
| zgc:158291 | 0.56 |
| rs1b | 0.55 |
| cabp2a | 0.54 |
| gnb1a | 0.54 |
Gene Ontology
Download
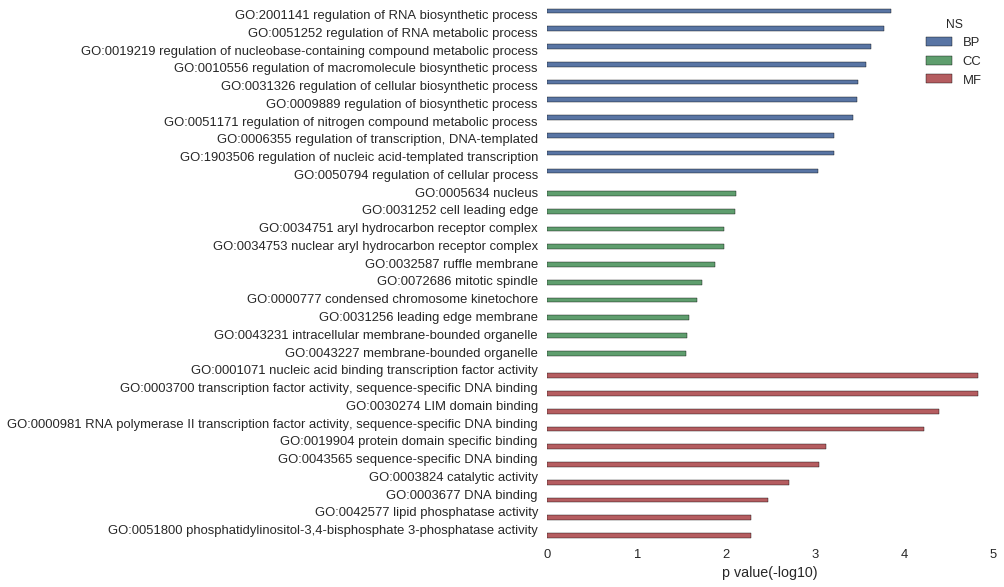
| GO | P value |
|---|---|
| GO:2001141 | 1.41e-04 |
| GO:0051252 | 1.69e-04 |
| GO:0019219 | 2.34e-04 |
| GO:0010556 | 2.67e-04 |
| GO:0031326 | 3.27e-04 |
| GO:0009889 | 3.37e-04 |
| GO:0051171 | 3.76e-04 |
| GO:0006355 | 6.06e-04 |
| GO:1903506 | 6.10e-04 |
| GO:0050794 | 9.25e-04 |
| GO:0005634 | 7.74e-03 |
| GO:0031252 | 7.87e-03 |
| GO:0034751 | 1.05e-02 |
| GO:0034753 | 1.05e-02 |
| GO:0032587 | 1.31e-02 |
| GO:0072686 | 1.83e-02 |
| GO:0000777 | 2.08e-02 |
| GO:0031256 | 2.60e-02 |
| GO:0043231 | 2.69e-02 |
| GO:0043227 | 2.77e-02 |
| GO:0001071 | 1.49e-05 |
| GO:0003700 | 1.49e-05 |
| GO:0030274 | 4.02e-05 |
| GO:0000981 | 6.00e-05 |
| GO:0019904 | 7.46e-04 |
| GO:0043565 | 9.10e-04 |
| GO:0003824 | 1.95e-03 |
| GO:0003677 | 3.31e-03 |
| GO:0042577 | 5.25e-03 |
| GO:0051800 | 5.25e-03 |
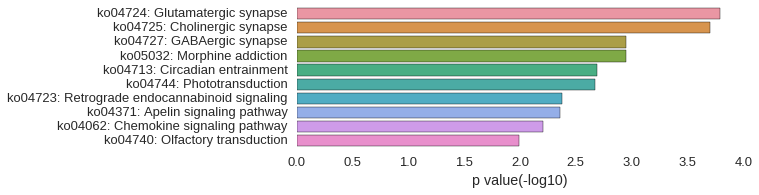
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08909
GGGAGTCTCCCATATTTTAGACTCATCTTCCGCCACCCTCACGTTTTGTTATTTCTTCCGGAAAACTCCC
GTAATTTCAACCATACCACCCTCCTGGTCCTCAGCATTTTGGTACGGTTTGTGAAACCCCTGGGTCGTCA
ACAATGGTATAGGATACGATTGCAGCGGCCGCCCAAATCACACATCATAGTACAGTTACTAACACCACAG
TACACAGTACCCAGTTCTCAACAGTCAACAGTGTTCTTCTGACACCTCACTCCCAAACCAAACCCTCAGG
TATCCCATATTTTCATGGATTAATGTTGGCAGGTATGCCATCAACC
GGGAGTCTCCCATATTTTAGACTCATCTTCCGCCACCCTCACGTTTTGTTATTTCTTCCGGAAAACTCCC
GTAATTTCAACCATACCACCCTCCTGGTCCTCAGCATTTTGGTACGGTTTGTGAAACCCCTGGGTCGTCA
ACAATGGTATAGGATACGATTGCAGCGGCCGCCCAAATCACACATCATAGTACAGTTACTAACACCACAG
TACACAGTACCCAGTTCTCAACAGTCAACAGTGTTCTTCTGACACCTCACTCCCAAACCAAACCCTCAGG
TATCCCATATTTTCATGGATTAATGTTGGCAGGTATGCCATCAACC