ZFLNCG08928
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023145 | heart | normal | 515.61 |
| ERR023144 | brain | normal | 446.07 |
| ERR023143 | swim bladder | normal | 182.44 |
| SRR592702 | pineal gland | normal | 168.56 |
| SRR594771 | endothelium | normal | 153.21 |
| SRR592703 | pineal gland | normal | 126.90 |
| SRR1569491 | embryo | normal | 117.99 |
| ERR023146 | head kidney | normal | 106.65 |
| SRR594769 | blood | normal | 104.57 |
| SRR592701 | pineal gland | normal | 98.52 |
Express in tissues
Correlated coding gene
Download
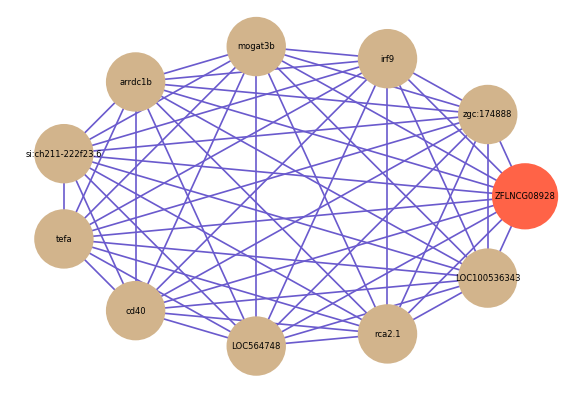
| gene | correlation coefficent |
|---|---|
| zgc:174888 | 0.77 |
| irf9 | 0.66 |
| mogat3b | 0.65 |
| arrdc1b | 0.65 |
| si:ch211-222f23.6 | 0.64 |
| tefa | 0.64 |
| cd40 | 0.63 |
| LOC564748 | 0.63 |
| rca2.1 | 0.63 |
| LOC100536343 | 0.62 |
Gene Ontology
Download
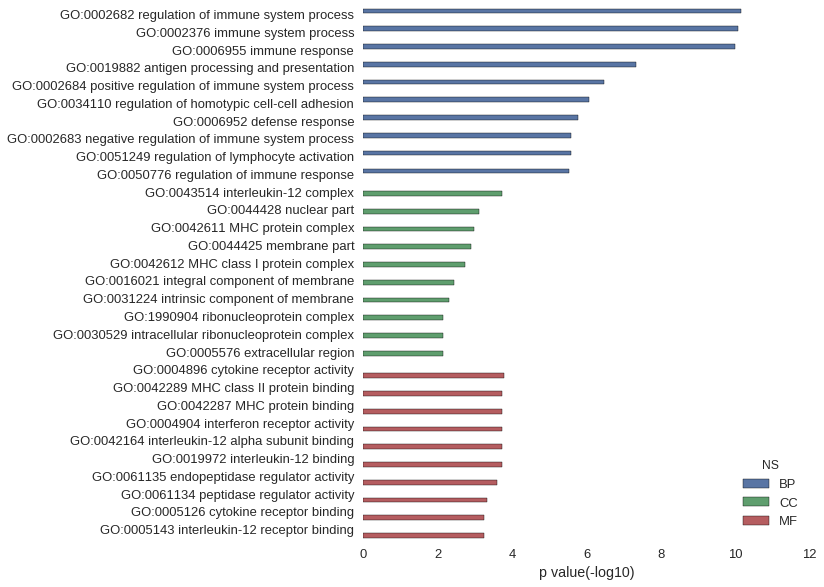
| GO | P value |
|---|---|
| GO:0002682 | 7.12e-11 |
| GO:0002376 | 8.58e-11 |
| GO:0006955 | 1.00e-10 |
| GO:0019882 | 4.64e-08 |
| GO:0002684 | 3.35e-07 |
| GO:0034110 | 8.78e-07 |
| GO:0006952 | 1.65e-06 |
| GO:0002683 | 2.62e-06 |
| GO:0051249 | 2.62e-06 |
| GO:0050776 | 3.02e-06 |
| GO:0043514 | 1.91e-04 |
| GO:0044428 | 7.86e-04 |
| GO:0042611 | 1.05e-03 |
| GO:0044425 | 1.31e-03 |
| GO:0042612 | 1.86e-03 |
| GO:0016021 | 3.76e-03 |
| GO:0031224 | 4.87e-03 |
| GO:1990904 | 7.14e-03 |
| GO:0030529 | 7.14e-03 |
| GO:0005576 | 7.35e-03 |
| GO:0004896 | 1.68e-04 |
| GO:0042289 | 1.91e-04 |
| GO:0042287 | 1.91e-04 |
| GO:0004904 | 1.91e-04 |
| GO:0042164 | 1.91e-04 |
| GO:0019972 | 1.91e-04 |
| GO:0061135 | 2.49e-04 |
| GO:0061134 | 4.76e-04 |
| GO:0005126 | 5.56e-04 |
| GO:0005143 | 5.68e-04 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08928
ATGTCTGCACTGATTGTATCTATCATTTCTGAATTGTGAATTTTGTGTCATTCGTAATAATTCAGGTAAA
ACCTGTCGACTGAATGTTTTAACAATGAATGTAGTGAAAAAGCGAACTATACATGAAGCCAAAAGTATCC
GCTTATCGCTCAGTGTTTAAGAGGACAACAAAACTTATTTACTACAGTAAATTCAAGCCATATGAAACTG
TAAAACTGTAGCATAGGAACCACAGCACCATCAGATGCAAGCTACTCTGTGAAATAAAAAGAAAAAAGTT
ACAAAAGTCATGTTTACAAGTATCCCTGTAGAGAAGGTTACTACTACAAACACGTTTTACTCAAACGAAT
TTAACAATCCGTCTGTTGTTGATGTACATGTGTTGTCTGTTTTTACCTGTAGATT
ATGTCTGCACTGATTGTATCTATCATTTCTGAATTGTGAATTTTGTGTCATTCGTAATAATTCAGGTAAA
ACCTGTCGACTGAATGTTTTAACAATGAATGTAGTGAAAAAGCGAACTATACATGAAGCCAAAAGTATCC
GCTTATCGCTCAGTGTTTAAGAGGACAACAAAACTTATTTACTACAGTAAATTCAAGCCATATGAAACTG
TAAAACTGTAGCATAGGAACCACAGCACCATCAGATGCAAGCTACTCTGTGAAATAAAAAGAAAAAAGTT
ACAAAAGTCATGTTTACAAGTATCCCTGTAGAGAAGGTTACTACTACAAACACGTTTTACTCAAACGAAT
TTAACAATCCGTCTGTTGTTGATGTACATGTGTTGTCTGTTTTTACCTGTAGATT