ZFLNCG08963
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562531 | muscle | normal | 4.11 |
| SRR1562529 | intestine and pancreas | normal | 3.52 |
| SRR891512 | blood | normal | 2.94 |
| SRR891495 | heart | normal | 2.88 |
| SRR1647682 | spleen | normal | 2.62 |
| SRR1562532 | spleen | normal | 2.06 |
| ERR023144 | brain | normal | 1.91 |
| SRR1562528 | eye | normal | 1.59 |
| SRR516122 | skin | male and 3.5 year | 1.48 |
| SRR1647681 | head kidney | SVCV treatment | 1.44 |
Express in tissues
Correlated coding gene
Download
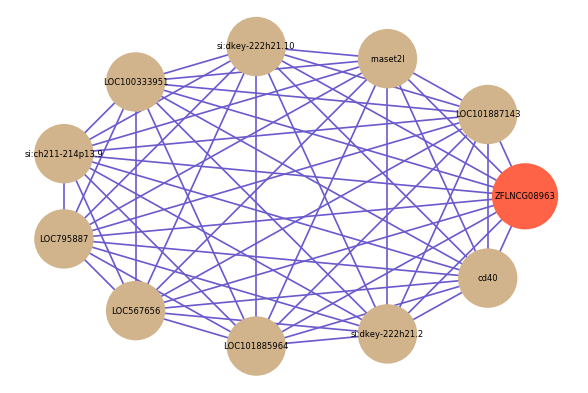
| gene | correlation coefficent |
|---|---|
| LOC101887143 | 0.61 |
| rnaset2l | 0.58 |
| si:dkey-222h21.10 | 0.58 |
| LOC100333951 | 0.57 |
| si:ch211-214p13.9 | 0.56 |
| LOC795887 | 0.56 |
| LOC567656 | 0.56 |
| LOC101885964 | 0.56 |
| si:dkey-222h21.2 | 0.56 |
| cd40 | 0.56 |
Gene Ontology
Download
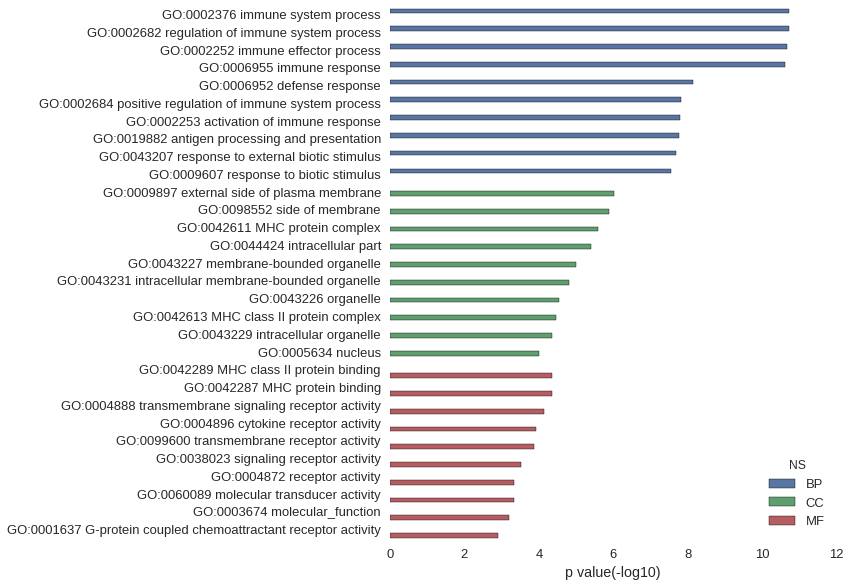
| GO | P value |
|---|---|
| GO:0002376 | 1.85e-11 |
| GO:0002682 | 1.88e-11 |
| GO:0002252 | 2.18e-11 |
| GO:0006955 | 2.44e-11 |
| GO:0006952 | 7.15e-09 |
| GO:0002684 | 1.50e-08 |
| GO:0002253 | 1.60e-08 |
| GO:0019882 | 1.67e-08 |
| GO:0043207 | 2.07e-08 |
| GO:0009607 | 2.75e-08 |
| GO:0009897 | 9.26e-07 |
| GO:0098552 | 1.33e-06 |
| GO:0042611 | 2.51e-06 |
| GO:0044424 | 3.94e-06 |
| GO:0043227 | 9.77e-06 |
| GO:0043231 | 1.50e-05 |
| GO:0043226 | 2.90e-05 |
| GO:0042613 | 3.46e-05 |
| GO:0043229 | 4.38e-05 |
| GO:0005634 | 9.93e-05 |
| GO:0042289 | 4.51e-05 |
| GO:0042287 | 4.51e-05 |
| GO:0004888 | 7.49e-05 |
| GO:0004896 | 1.21e-04 |
| GO:0099600 | 1.33e-04 |
| GO:0038023 | 3.05e-04 |
| GO:0004872 | 4.79e-04 |
| GO:0060089 | 4.79e-04 |
| GO:0003674 | 6.38e-04 |
| GO:0001637 | 1.23e-03 |
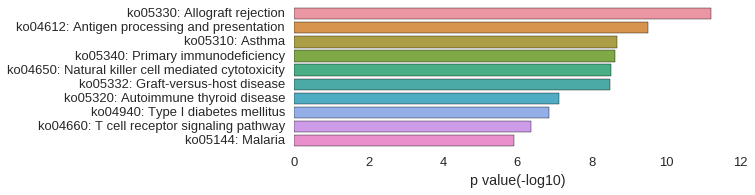
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG08963
GAAGAGCAACTCTTTATCCAGTCAGTGCACAGAGGAAGAAAAGGTCCCAATGCCAGTGCAGGAGATGTGT
GGGAAGACCGAGAAACTGGAGGATGTCTGAGAGAGGAGAAAGAAGACCACTGGTGCGTCTGAGTATTGAG
TATTTACTCTAGTACAAACCTGCTGCTGCCATTTACTCTGCACTGAGGTGGATCATCCAGATTGTTACGA
GAGAACAGAATGACGGTCCATCCTGAGAAATGCTGAGACGGAGAGAGATTTGAGCTCCCACTTCACATAA
AGACAAACACAACTTTTCACATACAGCTCA
GAAGAGCAACTCTTTATCCAGTCAGTGCACAGAGGAAGAAAAGGTCCCAATGCCAGTGCAGGAGATGTGT
GGGAAGACCGAGAAACTGGAGGATGTCTGAGAGAGGAGAAAGAAGACCACTGGTGCGTCTGAGTATTGAG
TATTTACTCTAGTACAAACCTGCTGCTGCCATTTACTCTGCACTGAGGTGGATCATCCAGATTGTTACGA
GAGAACAGAATGACGGTCCATCCTGAGAAATGCTGAGACGGAGAGAGATTTGAGCTCCCACTTCACATAA
AGACAAACACAACTTTTCACATACAGCTCA