ZFLNCG09096
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1648856 | brain | normal | 12.51 |
| ERR023144 | brain | normal | 10.18 |
| SRR1565813 | brain | normal | 4.09 |
| SRR1565819 | brain | normal | 3.40 |
| SRR630464 | 5 dpf | injected epidermidis | 3.25 |
| SRR1648854 | brain | normal | 3.20 |
| SRR1342219 | embryo | marco morpholino and infection with Mycobacterium marinum | 3.14 |
| SRR1049946 | embryo | transgenic TgOMP-Gal4 and UAS-GCaMP1.6 and treated with DMSO | 2.86 |
| SRR1565817 | brain | normal | 2.66 |
| SRR1291414 | 5 dpi | normal | 2.18 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
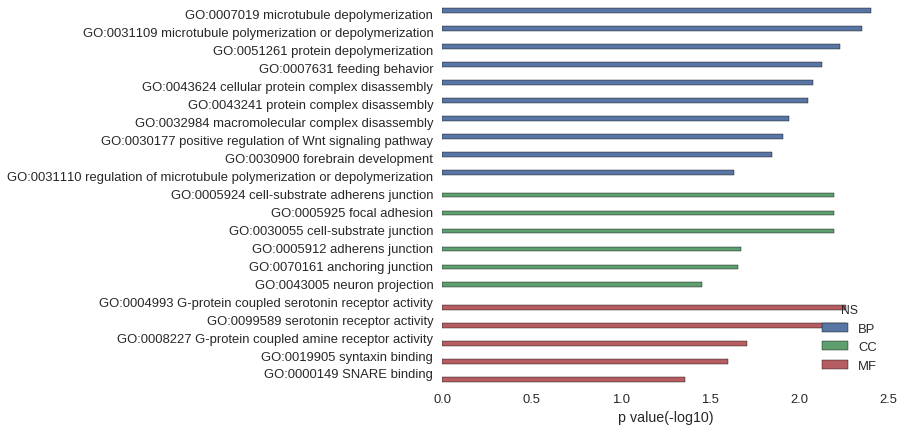
| GO | P value |
|---|---|
| GO:0007019 | 3.97e-03 |
| GO:0031109 | 4.47e-03 |
| GO:0051261 | 5.96e-03 |
| GO:0007631 | 7.44e-03 |
| GO:0043624 | 8.43e-03 |
| GO:0043241 | 8.92e-03 |
| GO:0032984 | 1.14e-02 |
| GO:0030177 | 1.24e-02 |
| GO:0030900 | 1.43e-02 |
| GO:0031110 | 2.32e-02 |
| GO:0005924 | 6.45e-03 |
| GO:0005925 | 6.45e-03 |
| GO:0030055 | 6.45e-03 |
| GO:0005912 | 2.12e-02 |
| GO:0070161 | 2.22e-02 |
| GO:0043005 | 3.53e-02 |
| GO:0004993 | 5.46e-03 |
| GO:0099589 | 5.46e-03 |
| GO:0008227 | 1.97e-02 |
| GO:0019905 | 2.51e-02 |
| GO:0000149 | 4.39e-02 |
KEGG Pathway
KEGG pathway of this lncRNA gene hava does not exist!
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09096
CGAGTATTCAAAGGGTATTTAAAAGAACTAGGGGAAATATCCTTGAACAGTCTTAAACTAAACTAAATGT
ACATGAAAATTCTGAATAAATTTTAGATTTGAAGAGAGATTTTGAGATTTTTTTTTTTTGTTGTTGTTGT
TTGTTTGTTTTTAGAATATGCAAATTGGCATTATTTGATGCAGAAATACAGCATGTGATTGAGCTATTGT
ATCTCCTAATGGATTTAGCAGTGTCCACTACTTTGGGTGCAGATGTCCCATTTTAAAGACTTTTTTTCTG
TGTTGTTTGGTAGGGTAGAGTGAGTTTGGACTTGCTCAGCTGTAGTGTGGACATGCAATAATGTTTTTTT
TTTCAATCTATTTAAAGTGTAA
CGAGTATTCAAAGGGTATTTAAAAGAACTAGGGGAAATATCCTTGAACAGTCTTAAACTAAACTAAATGT
ACATGAAAATTCTGAATAAATTTTAGATTTGAAGAGAGATTTTGAGATTTTTTTTTTTTGTTGTTGTTGT
TTGTTTGTTTTTAGAATATGCAAATTGGCATTATTTGATGCAGAAATACAGCATGTGATTGAGCTATTGT
ATCTCCTAATGGATTTAGCAGTGTCCACTACTTTGGGTGCAGATGTCCCATTTTAAAGACTTTTTTTCTG
TGTTGTTTGGTAGGGTAGAGTGAGTTTGGACTTGCTCAGCTGTAGTGTGGACATGCAATAATGTTTTTTT
TTTCAATCTATTTAAAGTGTAA