ZFLNCG09116
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800049 | sphere stage | control treatment | 11.63 |
| SRR535978 | larvae | normal | 9.87 |
| SRR800046 | sphere stage | 5azaCyD treatment | 9.57 |
| SRR658541 | 6 somite | normal | 9.30 |
| SRR516124 | skin | male and 3.5 year | 8.78 |
| SRR1647679 | head kidney | normal | 8.63 |
| SRR658543 | 6 somite | Gata5 morphan | 8.49 |
| SRR1035981 | 13 hpf | rx3+/+ or rx3+/- | 8.48 |
| SRR1035978 | 13 hpf | rx3-/- | 7.70 |
| SRR1647684 | spleen | SVCV treatment | 7.54 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| zgc:174651 | 0.58 |
| zgc:171551 | 0.57 |
| lamtor1 | 0.57 |
| LOC101886556 | 0.55 |
| trim35-39 | 0.54 |
| nr5a1a | 0.53 |
| supt4h1 | 0.53 |
| si:dkey-20i20.2 | 0.52 |
| helq | 0.51 |
| zgc:113176 | 0.50 |
Gene Ontology
Download
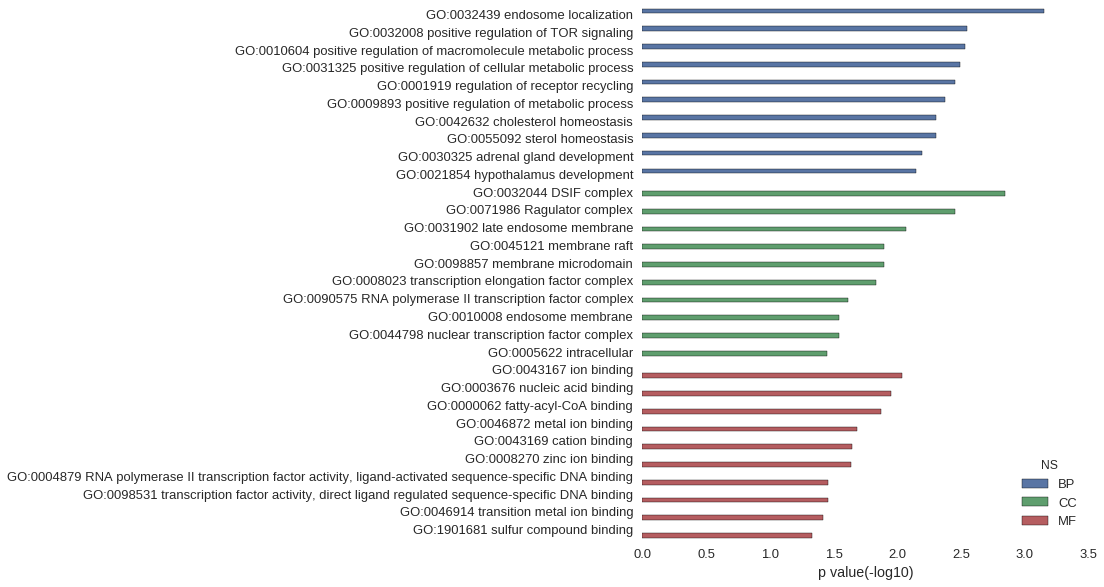
| GO | P value |
|---|---|
| GO:0032439 | 7.11e-04 |
| GO:0032008 | 2.84e-03 |
| GO:0010604 | 2.95e-03 |
| GO:0031325 | 3.23e-03 |
| GO:0001919 | 3.55e-03 |
| GO:0009893 | 4.27e-03 |
| GO:0042632 | 4.96e-03 |
| GO:0055092 | 4.96e-03 |
| GO:0030325 | 6.38e-03 |
| GO:0021854 | 7.09e-03 |
| GO:0032044 | 1.42e-03 |
| GO:0071986 | 3.55e-03 |
| GO:0031902 | 8.50e-03 |
| GO:0045121 | 1.27e-02 |
| GO:0098857 | 1.27e-02 |
| GO:0008023 | 1.48e-02 |
| GO:0090575 | 2.46e-02 |
| GO:0010008 | 2.88e-02 |
| GO:0044798 | 2.88e-02 |
| GO:0005622 | 3.58e-02 |
| GO:0043167 | 9.23e-03 |
| GO:0003676 | 1.12e-02 |
| GO:0000062 | 1.34e-02 |
| GO:0046872 | 2.09e-02 |
| GO:0043169 | 2.27e-02 |
| GO:0008270 | 2.32e-02 |
| GO:0004879 | 3.50e-02 |
| GO:0098531 | 3.50e-02 |
| GO:0046914 | 3.81e-02 |
| GO:1901681 | 4.66e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09116
TGAGAACACACAAGACAACTGTATCTACAGTGGACGTTAACACACACACACACACACACACACACACACA
CACACACACACACACACACACACACACACACACACATTTACATTTCACTGATATTAATCAAATGAATACT
ATTACTGAGCTCTTGCAACTATCCAAATAATCACCTTCCGTTTTTCATCAGAGTAGCTTCATCATCTGGT
ATAAAGTTTGACAAAAGATCTGAAACACGTCGTTTCTATTCAAGGACAAGAAACAGACAGTAAGACAGTA
TTCGGCTTGTGCATGTAATACAGACACATCTTCTGACACTTTTATTATAACGTTACCTTCAGTATATTAA
GCTGGCTTTGATCATCCCCTTCTCTTTTGAAAGACATGCTTGGCGACCTATTCTTCCAGAAACTAAACAG
TGTTTTCTTATTTTGCTGCCAAAAGCTTACAAACTCCTCTCTTGTACTTCCTGTTTCTATGTGGAATTAT
GGGTAGTGATGTCC
TGAGAACACACAAGACAACTGTATCTACAGTGGACGTTAACACACACACACACACACACACACACACACA
CACACACACACACACACACACACACACACACACACATTTACATTTCACTGATATTAATCAAATGAATACT
ATTACTGAGCTCTTGCAACTATCCAAATAATCACCTTCCGTTTTTCATCAGAGTAGCTTCATCATCTGGT
ATAAAGTTTGACAAAAGATCTGAAACACGTCGTTTCTATTCAAGGACAAGAAACAGACAGTAAGACAGTA
TTCGGCTTGTGCATGTAATACAGACACATCTTCTGACACTTTTATTATAACGTTACCTTCAGTATATTAA
GCTGGCTTTGATCATCCCCTTCTCTTTTGAAAGACATGCTTGGCGACCTATTCTTCCAGAAACTAAACAG
TGTTTTCTTATTTTGCTGCCAAAAGCTTACAAACTCCTCTCTTGTACTTCCTGTTTCTATGTGGAATTAT
GGGTAGTGATGTCC

