ZFLNCG09383
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1035239 | liver | transgenic mCherry control | 99.26 |
| SRR1035237 | liver | transgenic UHRF1 | 40.77 |
| SRR891504 | liver | normal | 29.30 |
| ERR023146 | head kidney | normal | 24.50 |
| SRR1291414 | 5 dpi | normal | 13.06 |
| SRR527835 | tail | normal | 12.30 |
| SRR1167756 | embryo | mpeg1 morpholino | 11.95 |
| SRR1562529 | intestine and pancreas | normal | 11.28 |
| SRR1028002 | head | normal | 10.51 |
| SRR1647679 | head kidney | normal | 9.41 |
Express in tissues
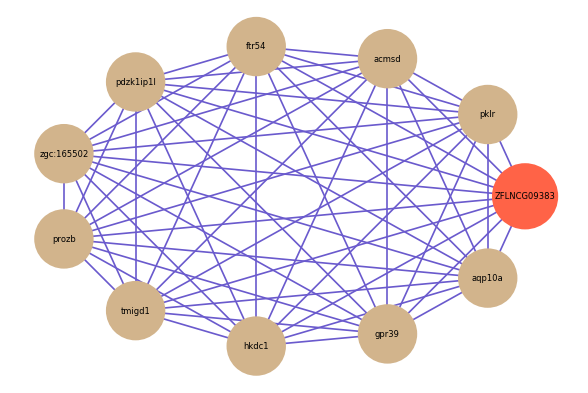
Correlated coding gene
Download
Gene Ontology
Download

| GO | P value |
|---|---|
| GO:0050878 | 7.98e-11 |
| GO:0044260 | 1.55e-09 |
| GO:0050817 | 2.47e-09 |
| GO:0007599 | 2.47e-09 |
| GO:0007596 | 2.47e-09 |
| GO:1900047 | 2.42e-08 |
| GO:0030195 | 2.42e-08 |
| GO:0061045 | 2.42e-08 |
| GO:0030193 | 7.13e-08 |
| GO:1900046 | 7.13e-08 |
| GO:0005615 | 1.70e-09 |
| GO:0044421 | 1.20e-08 |
| GO:0044424 | 1.42e-07 |
| GO:0005576 | 5.05e-07 |
| GO:0043229 | 7.43e-07 |
| GO:0043226 | 9.39e-07 |
| GO:0044422 | 3.58e-06 |
| GO:0044446 | 5.26e-06 |
| GO:0032991 | 5.28e-06 |
| GO:0044464 | 1.97e-05 |
| GO:0030414 | 7.94e-12 |
| GO:0004857 | 1.21e-11 |
| GO:0061134 | 2.55e-11 |
| GO:0004866 | 4.26e-11 |
| GO:0061135 | 8.97e-11 |
| GO:0004867 | 4.13e-10 |
| GO:0005215 | 6.74e-09 |
| GO:0008509 | 8.71e-08 |
| GO:0030234 | 2.15e-06 |
| GO:0008514 | 2.51e-06 |
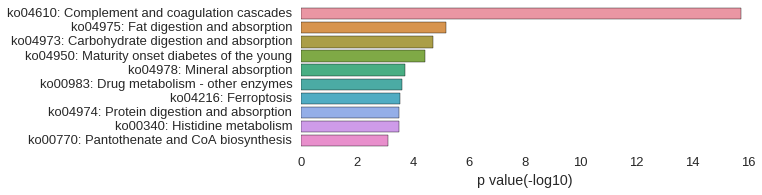
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09383
CACACACACAGATGGACAGTGAAGATGAGACCGTCATGTTGGGTTTGGATTACTGCACTGAATCGTGTTT
ATTATCCGTCTCTGCATCTTCATTTGCATAAATGTACTGGCTAAATGCAAATGAGCTCAGTGTGCGAAAT
TAACTTCTCTTAGCAGTGTGTGTGTGTGTGTGTGTGTCTGACCGATGGTGCAGATGATGCTGGTGTTGCG
GGCAGTGTGTGTGTGTGTGTGTGTGTGTCTCTGACCCATCGTGCACATGATGCTGGTGTTGCGGGCAGTG
ATGCGGCATCCTGGTCGATGTCCAGCAGACCCAGATGCTCCACGAACGTGTCGGCCATCGATGCGTCCAG
CTGCTGCTTCTGGATGAATGAGTCCGGCAGAGCCATGACCTCTGAGTAACGCCGGATACGAGCTGAATGA
TACACACATAATGAAACACACACAAAGATAAAGAGAGAAACACACACACACACANNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNACACACACACACACACACACACACACACACACATGAGAGAAACACAAATACAACAC
AATGAGACACACACACACACACACACACACACACACTGCCTGTATTATCTACACTATTAGCCTCTCTCAC
ACACACATGCTGGTATAGGTGGTTTATGAGGACTCTCCACAGGTGTAATGTATTTTATATGGTAATTCTC
TTATTATAAGCTCCTCCCCCTAACCCCACCCACACACACCTGTGGACAACCTGAGTGCTCAAATTATTTT
TAAGCATTCGTAACTATCAGGACCTCAGACATGTCCTCAAACCAGCTCAGTGCTGTAATACTTAGATCAT
ACCCACAACATTATACACATTTCTGTCCTCATAAATCAACACACACACACACACACACACACACACACAC
ACACACACACCTGATGAGTGCCATGAGAGAAGTTGAGTCTGGCGATGTTCATTCCTGCTTTCACCATCTC
CTGTAGTTTAGTGATGGATCGAGACGCTGGACCTGAAACACACACACACACACACACACACACACACACA
CACACACAAACAAACAAACACATTAGTGATCTAAAACACTTTAAAATCAGTAACAACATGCTGGTGTTGC
GGGCAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTATGTGTCTGACCGATGGTGCAGATGATGCTGGTGT
TGCGGGCAGTGTGTGTGTGTGTGTGTGTGTGTGT
CACACACACAGATGGACAGTGAAGATGAGACCGTCATGTTGGGTTTGGATTACTGCACTGAATCGTGTTT
ATTATCCGTCTCTGCATCTTCATTTGCATAAATGTACTGGCTAAATGCAAATGAGCTCAGTGTGCGAAAT
TAACTTCTCTTAGCAGTGTGTGTGTGTGTGTGTGTGTCTGACCGATGGTGCAGATGATGCTGGTGTTGCG
GGCAGTGTGTGTGTGTGTGTGTGTGTGTCTCTGACCCATCGTGCACATGATGCTGGTGTTGCGGGCAGTG
ATGCGGCATCCTGGTCGATGTCCAGCAGACCCAGATGCTCCACGAACGTGTCGGCCATCGATGCGTCCAG
CTGCTGCTTCTGGATGAATGAGTCCGGCAGAGCCATGACCTCTGAGTAACGCCGGATACGAGCTGAATGA
TACACACATAATGAAACACACACAAAGATAAAGAGAGAAACACACACACACACANNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNACACACACACACACACACACACACACACACACATGAGAGAAACACAAATACAACAC
AATGAGACACACACACACACACACACACACACACACTGCCTGTATTATCTACACTATTAGCCTCTCTCAC
ACACACATGCTGGTATAGGTGGTTTATGAGGACTCTCCACAGGTGTAATGTATTTTATATGGTAATTCTC
TTATTATAAGCTCCTCCCCCTAACCCCACCCACACACACCTGTGGACAACCTGAGTGCTCAAATTATTTT
TAAGCATTCGTAACTATCAGGACCTCAGACATGTCCTCAAACCAGCTCAGTGCTGTAATACTTAGATCAT
ACCCACAACATTATACACATTTCTGTCCTCATAAATCAACACACACACACACACACACACACACACACAC
ACACACACACCTGATGAGTGCCATGAGAGAAGTTGAGTCTGGCGATGTTCATTCCTGCTTTCACCATCTC
CTGTAGTTTAGTGATGGATCGAGACGCTGGACCTGAAACACACACACACACACACACACACACACACACA
CACACACAAACAAACAAACACATTAGTGATCTAAAACACTTTAAAATCAGTAACAACATGCTGGTGTTGC
GGGCAGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTATGTGTCTGACCGATGGTGCAGATGATGCTGGTGT
TGCGGGCAGTGTGTGTGTGTGTGTGTGTGTGTGT