ZFLNCG09460
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR592703 | pineal gland | normal | 96.12 |
| SRR592702 | pineal gland | normal | 49.93 |
| SRR592699 | pineal gland | normal | 21.71 |
| SRR519742 | 4 dpf | VD3 treatment | 20.42 |
| SRR519737 | 2 dpf | VD3 treatment | 19.34 |
| SRR516135 | skin | male and 5 month | 19.04 |
| SRR519722 | 4 dpf | FETOH treatment | 16.95 |
| SRR592701 | pineal gland | normal | 14.76 |
| SRR519727 | 6 dpf | FETOH treatment | 14.75 |
| SRR519717 | 2 dpf | FETOH treatment | 13.85 |
Express in tissues
Correlated coding gene
Download
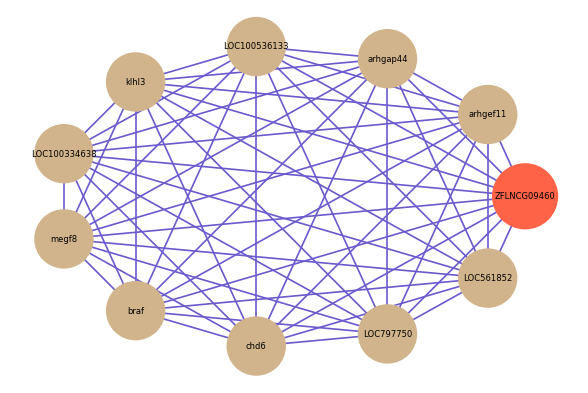
| gene | correlation coefficent |
|---|---|
| arhgef11 | 0.59 |
| arhgap44 | 0.57 |
| LOC100536133 | 0.55 |
| klhl3 | 0.55 |
| LOC100334638 | 0.54 |
| megf8 | 0.54 |
| braf | 0.54 |
| chd6 | 0.53 |
| LOC797750 | 0.53 |
| LOC561852 | 0.53 |
Gene Ontology
Download
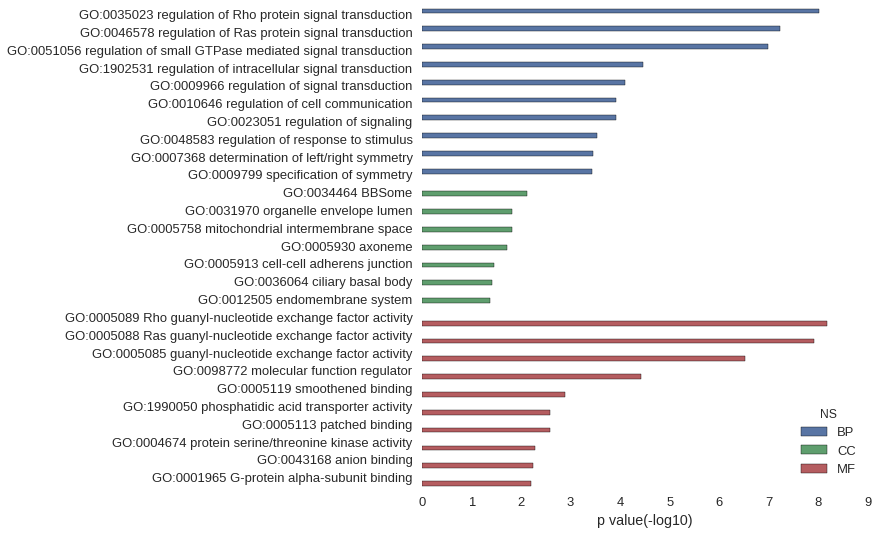
| GO | P value |
|---|---|
| GO:0035023 | 9.76e-09 |
| GO:0046578 | 6.10e-08 |
| GO:0051056 | 1.06e-07 |
| GO:1902531 | 3.45e-05 |
| GO:0009966 | 7.90e-05 |
| GO:0010646 | 1.22e-04 |
| GO:0023051 | 1.23e-04 |
| GO:0048583 | 2.89e-04 |
| GO:0007368 | 3.58e-04 |
| GO:0009799 | 3.67e-04 |
| GO:0034464 | 7.65e-03 |
| GO:0031970 | 1.52e-02 |
| GO:0005758 | 1.52e-02 |
| GO:0005930 | 1.90e-02 |
| GO:0005913 | 3.52e-02 |
| GO:0036064 | 3.89e-02 |
| GO:0012505 | 4.26e-02 |
| GO:0005089 | 6.85e-09 |
| GO:0005088 | 1.25e-08 |
| GO:0005085 | 2.99e-07 |
| GO:0098772 | 3.77e-05 |
| GO:0005119 | 1.28e-03 |
| GO:1990050 | 2.56e-03 |
| GO:0005113 | 2.56e-03 |
| GO:0004674 | 5.15e-03 |
| GO:0043168 | 5.76e-03 |
| GO:0001965 | 6.38e-03 |
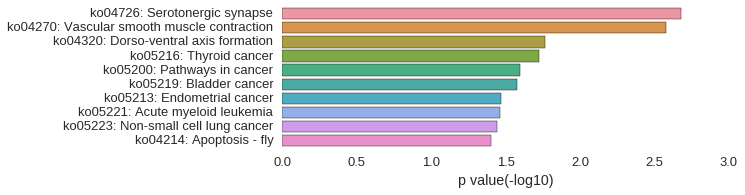
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09460
CAGTCTACAGAGATTTCCCAGTATTTCTCTGTTACTATCACGTTATCACATAAATTAACCCTGAAAAAGC
CAAAGCAAAGCAAACACGCTTGACAGTGTGTCAAAAAGAGAAGACCTGAAAGAAGGATGTGTATATTTCT
ATGCAGACTCTGAGCCTACCGAGCCGCTGAGTGTAGACGGAAACTCATCAGAACTGGATATGGAGGATCT
GGATGAGGAAGAGGAAGAGG
CAGTCTACAGAGATTTCCCAGTATTTCTCTGTTACTATCACGTTATCACATAAATTAACCCTGAAAAAGC
CAAAGCAAAGCAAACACGCTTGACAGTGTGTCAAAAAGAGAAGACCTGAAAGAAGGATGTGTATATTTCT
ATGCAGACTCTGAGCCTACCGAGCCGCTGAGTGTAGACGGAAACTCATCAGAACTGGATATGGAGGATCT
GGATGAGGAAGAGGAAGAGG