ZFLNCG09466
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR514028 | retina | control morpholino | 50.76 |
| SRR527833 | 5 dpf | normal | 44.32 |
| SRR372796 | bud | normal | 38.23 |
| SRR535978 | larvae | normal | 34.38 |
| SRR372800 | 2 dpf | normal | 33.94 |
| SRR527834 | head | normal | 33.02 |
| SRR514030 | retina | id2a morpholino | 31.03 |
| SRR372798 | 28 hpf | normal | 25.94 |
| SRR527836 | 24hpf | normal | 25.77 |
| SRR1035237 | liver | transgenic UHRF1 | 24.01 |
Express in tissues
Correlated coding gene
Download
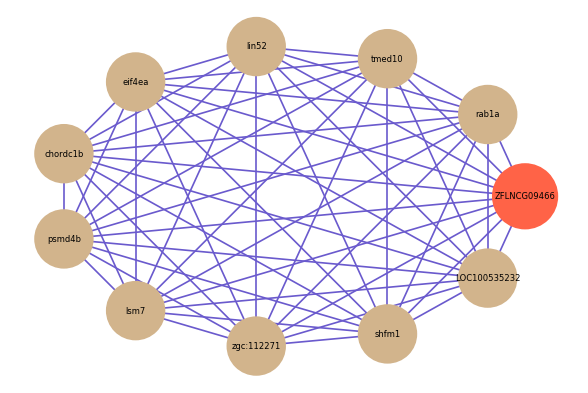
| gene | correlation coefficent |
|---|---|
| rab1a | 0.66 |
| tmed10 | 0.65 |
| lin52 | 0.64 |
| eif4ea | 0.63 |
| chordc1b | 0.63 |
| psmd4b | 0.63 |
| lsm7 | 0.63 |
| zgc:112271 | 0.63 |
| shfm1 | 0.62 |
| LOC100535232 | 0.61 |
Gene Ontology
Download
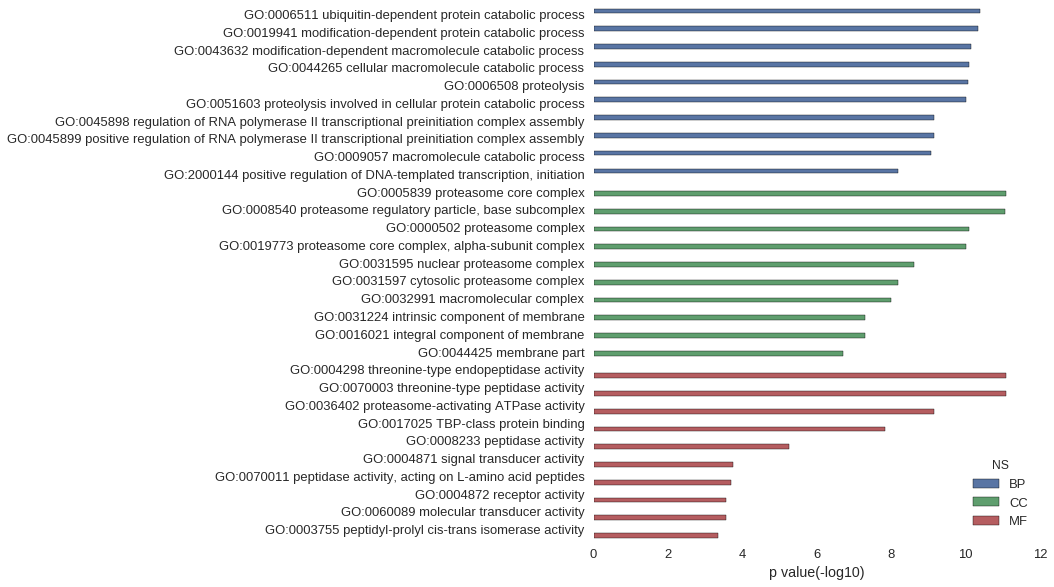
| GO | P value |
|---|---|
| GO:0006511 | 4.13e-11 |
| GO:0019941 | 4.55e-11 |
| GO:0043632 | 7.29e-11 |
| GO:0044265 | 8.05e-11 |
| GO:0006508 | 8.39e-11 |
| GO:0051603 | 9.78e-11 |
| GO:0045898 | 6.91e-10 |
| GO:0045899 | 6.91e-10 |
| GO:0009057 | 8.62e-10 |
| GO:2000144 | 6.35e-09 |
| GO:0005839 | 8.03e-12 |
| GO:0008540 | 8.92e-12 |
| GO:0000502 | 8.17e-11 |
| GO:0019773 | 9.60e-11 |
| GO:0031595 | 2.40e-09 |
| GO:0031597 | 6.35e-09 |
| GO:0032991 | 1.04e-08 |
| GO:0031224 | 4.93e-08 |
| GO:0016021 | 5.02e-08 |
| GO:0044425 | 1.94e-07 |
| GO:0004298 | 8.03e-12 |
| GO:0070003 | 8.03e-12 |
| GO:0036402 | 6.91e-10 |
| GO:0017025 | 1.42e-08 |
| GO:0008233 | 5.70e-06 |
| GO:0004871 | 1.76e-04 |
| GO:0070011 | 2.04e-04 |
| GO:0004872 | 2.82e-04 |
| GO:0060089 | 2.82e-04 |
| GO:0003755 | 4.65e-04 |
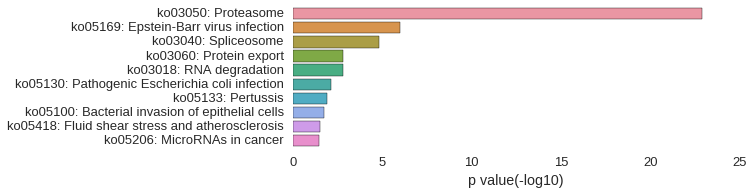
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09466
TCACTTTTATAAATCATATTTATTGGGAAGACTGAAATCTGCAAAAGCAAAACATTAGGACTACATTAAA
CATAAAAAACGTTTTTATTGTCTGCACAGTAAAGTTGAGAAATGGTCATAATGTGTGAAACATGGTATAC
ACAATCAGTGAATAGGAAAGTCAGGTTTGCAGCGCTGTGTTTACATGTTGTCCTCATCAGAGTCATCCTC
CTCCTCCAGGGATTCCTGTCAGGACAAATGTGACAATCAGCCGATAATCAAAAGAGCTTTTGTTGCAAAC
ATTACCAGAGAAAGTGACTTACTTTAGCGTATTTCTCAGCCTCCTCTTTGACATCTTTGGGGACGAGCTC
GTGCCTTCCTTTTGTGACTGATAACGGGTGGAAAAAAAGATATATAAATAAAATACTAAAGCTCTTTTCT
AGTGTAGACACACCATAAAATGTAAAACCCCTTTTAAAATTGCTGAATTACATTATGCCGATATCGGAAA
GATACCAGAATAAGTCATTTCCACTCCAGCATAAAAAAAACAACAACAACAACGAAAAAGGAGAAGTAAT
GAAC
TCACTTTTATAAATCATATTTATTGGGAAGACTGAAATCTGCAAAAGCAAAACATTAGGACTACATTAAA
CATAAAAAACGTTTTTATTGTCTGCACAGTAAAGTTGAGAAATGGTCATAATGTGTGAAACATGGTATAC
ACAATCAGTGAATAGGAAAGTCAGGTTTGCAGCGCTGTGTTTACATGTTGTCCTCATCAGAGTCATCCTC
CTCCTCCAGGGATTCCTGTCAGGACAAATGTGACAATCAGCCGATAATCAAAAGAGCTTTTGTTGCAAAC
ATTACCAGAGAAAGTGACTTACTTTAGCGTATTTCTCAGCCTCCTCTTTGACATCTTTGGGGACGAGCTC
GTGCCTTCCTTTTGTGACTGATAACGGGTGGAAAAAAAGATATATAAATAAAATACTAAAGCTCTTTTCT
AGTGTAGACACACCATAAAATGTAAAACCCCTTTTAAAATTGCTGAATTACATTATGCCGATATCGGAAA
GATACCAGAATAAGTCATTTCCACTCCAGCATAAAAAAAACAACAACAACAACGAAAAAGGAGAAGTAAT
GAAC