ZFLNCG09474
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023144 | brain | normal | 28.13 |
| SRR941749 | anterior pectoral fin | normal | 26.39 |
| SRR800037 | egg | normal | 22.01 |
| SRR941753 | posterior pectoral fin | normal | 19.36 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 18.90 |
| SRR1205154 | 5dpf | transgenic sqET20 | 18.30 |
| SRR1205151 | 5dpf | transgenic sqET20 and neomycin treated 1h | 18.27 |
| ERR023143 | swim bladder | normal | 17.11 |
| ERR594416 | retina | transgenic flk1 | 15.77 |
| SRR1647682 | spleen | normal | 15.76 |
Express in tissues
Correlated coding gene
Download
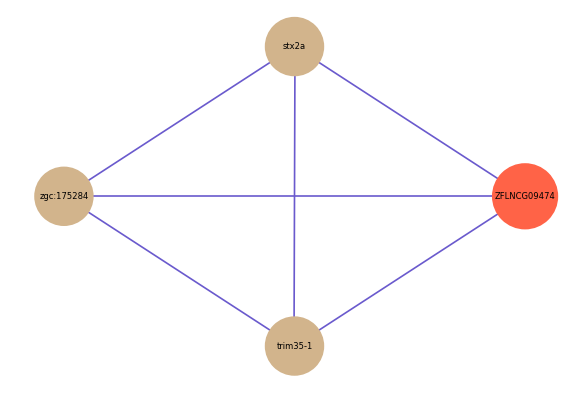
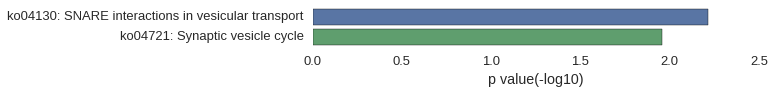
| gene | correlation coefficent |
|---|---|
| stx2a | 0.53 |
| zgc:175284 | 0.51 |
| trim35-1 | 0.50 |
Gene Ontology
Download
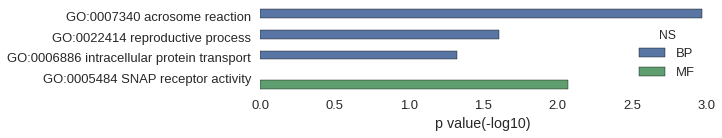
| GO | P value |
|---|---|
| GO:0007340 | 1.07e-03 |
| GO:0022414 | 2.47e-02 |
| GO:0006886 | 4.74e-02 |
| GO:0005484 | 8.50e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09474
TAAATACTGAGCAAAGGGTCTGAATACTGAGGACTATGTGATATTGGGTGCTCTTTGTACATTAGTGAGG
AAAAACAAGAAGTTAAATGATTTTAGCAAATGGCTGCACATATAACAAAGAGTAAAAAAGTTAAGGTGGT
CTGAATACTTTCTGTACCCACTGAAAGGATATCTGATACAACAACAGGCTTTTTCTTTTTGTCTGGACTG
AAGGGATCTTTCTTCTTGTTTTTTCTTGATTGTAACTCATCATTCCAGAGCTCTGTATGGGAAATATGAA
AATGCAGCATAAATAAGAGATAAATAAAAACATTATTTGTGATGGTGTATCAACAAATACCTGAGGTAAT
ATCTATACTATGCCGATCTTCCTCCAAACGTCTGATCTTCTCCTCTAATTCAGTCTGAACTGTGTCAAAT
AATAGAAGTTTCTCACTCTGGAAGATTGAAAAAAAAAATGATTAAATTGGACTGACTTGCATTGTTTAAT
CTTTAAGGTGAATTTAAATGTTAAAAGTGACAAATGGCTTTTAGAGGTATGCTACAACAGCTTGAAGCAA
TATCAACACTTCAGTGTGTATGGCTTTTAAACACGCACCATTGTTTCGGTAATAGTGGATTAAATCTGTA
TATCTCACCTCCCAGTGCTGGAACGCTGCTTGGGTTTCACAGTCATATTT
TAAATACTGAGCAAAGGGTCTGAATACTGAGGACTATGTGATATTGGGTGCTCTTTGTACATTAGTGAGG
AAAAACAAGAAGTTAAATGATTTTAGCAAATGGCTGCACATATAACAAAGAGTAAAAAAGTTAAGGTGGT
CTGAATACTTTCTGTACCCACTGAAAGGATATCTGATACAACAACAGGCTTTTTCTTTTTGTCTGGACTG
AAGGGATCTTTCTTCTTGTTTTTTCTTGATTGTAACTCATCATTCCAGAGCTCTGTATGGGAAATATGAA
AATGCAGCATAAATAAGAGATAAATAAAAACATTATTTGTGATGGTGTATCAACAAATACCTGAGGTAAT
ATCTATACTATGCCGATCTTCCTCCAAACGTCTGATCTTCTCCTCTAATTCAGTCTGAACTGTGTCAAAT
AATAGAAGTTTCTCACTCTGGAAGATTGAAAAAAAAAATGATTAAATTGGACTGACTTGCATTGTTTAAT
CTTTAAGGTGAATTTAAATGTTAAAAGTGACAAATGGCTTTTAGAGGTATGCTACAACAGCTTGAAGCAA
TATCAACACTTCAGTGTGTATGGCTTTTAAACACGCACCATTGTTTCGGTAATAGTGGATTAAATCTGTA
TATCTCACCTCCCAGTGCTGGAACGCTGCTTGGGTTTCACAGTCATATTT