ZFLNCG09496
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR726541 | 5 dpf | infection with Mycobacterium marinum | 11.09 |
| SRR038626 | embryo | control morpholino and bacterial infection | 10.95 |
| SRR941749 | anterior pectoral fin | normal | 10.15 |
| SRR527834 | head | normal | 9.71 |
| SRR065197 | 3 dpf | U1C knockout | 9.17 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 6.97 |
| SRR957181 | heart | normal | 6.39 |
| SRR594769 | blood | normal | 6.32 |
| SRR941753 | posterior pectoral fin | normal | 6.01 |
| SRR1188156 | embryo | Control PBS 4 hpi | 5.55 |
Express in tissues
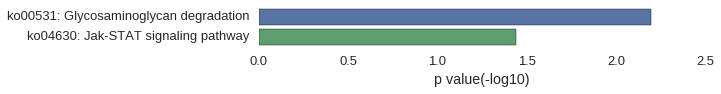
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| si:dkey-159f12.2 | 0.58 |
| crfb9 | 0.56 |
| zgc:64065 | 0.55 |
| nr5a1a | 0.54 |
| zgc:171551 | 0.54 |
| si:dkey-20i20.2 | 0.53 |
| LOC101883108 | 0.52 |
| zgc:136804 | 0.52 |
| zgc:113176 | 0.51 |
| v-fbpl | 0.51 |
Gene Ontology
Download
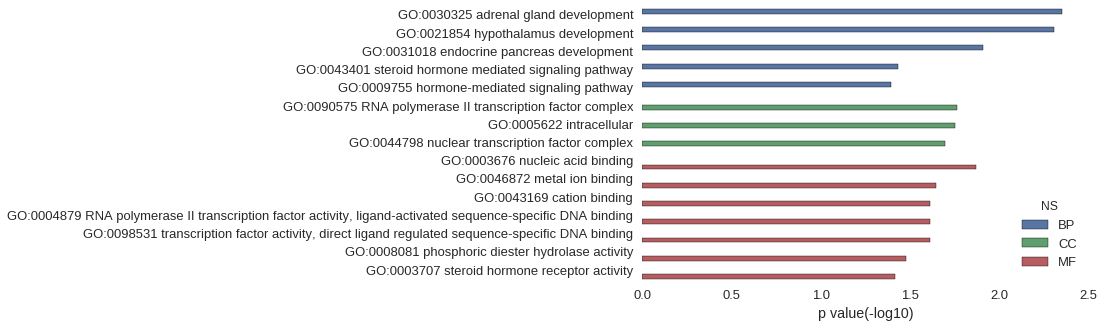
| GO | P value |
|---|---|
| GO:0030325 | 4.47e-03 |
| GO:0021854 | 4.96e-03 |
| GO:0031018 | 1.24e-02 |
| GO:0043401 | 3.72e-02 |
| GO:0009755 | 4.06e-02 |
| GO:0090575 | 1.73e-02 |
| GO:0005622 | 1.77e-02 |
| GO:0044798 | 2.02e-02 |
| GO:0003676 | 1.35e-02 |
| GO:0046872 | 2.27e-02 |
| GO:0043169 | 2.44e-02 |
| GO:0004879 | 2.46e-02 |
| GO:0098531 | 2.46e-02 |
| GO:0008081 | 3.33e-02 |
| GO:0003707 | 3.82e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09496
ACGATCTGCTCCCCAACTCCGTTCTCTATTTTTGTGCTTCCTTGTGCTGCCAAGCCCCTGACGTAATGCT
CCGTGTTAATTATTAGCATGTGGGAAGCGATGCCGAGACTCTCCTCTGCTGCTTAAACAAGAGCGACTGA
CTGCGGGGAGGAAAACCTCGTCAAGATGGCCAAACACTCTGACCCAGAGTGAAGGGGCACCGCTGCTCCC
TGTCCCGTCCCGTCCAGTCCTGTCCTACGACCATCACCAATGCAACTGCG
ACGATCTGCTCCCCAACTCCGTTCTCTATTTTTGTGCTTCCTTGTGCTGCCAAGCCCCTGACGTAATGCT
CCGTGTTAATTATTAGCATGTGGGAAGCGATGCCGAGACTCTCCTCTGCTGCTTAAACAAGAGCGACTGA
CTGCGGGGAGGAAAACCTCGTCAAGATGGCCAAACACTCTGACCCAGAGTGAAGGGGCACCGCTGCTCCC
TGTCCCGTCCCGTCCAGTCCTGTCCTACGACCATCACCAATGCAACTGCG