ZFLNCG09610
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 3.07 |
| SRR594771 | endothelium | normal | 2.83 |
| SRR749514 | 24 hpf | normal | 2.43 |
| ERR594417 | retina | normal | 2.33 |
| SRR516133 | skin | male and 5 month | 2.31 |
| SRR800045 | muscle | normal | 1.98 |
| SRR658547 | 6 somite | Gata5/6 morphant | 1.95 |
| SRR749515 | 24 hpf | eif3ha morphant | 1.78 |
| SRR658545 | 6 somite | Gata6 morphant | 1.73 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 1.73 |
Express in tissues
Correlated coding gene
Download
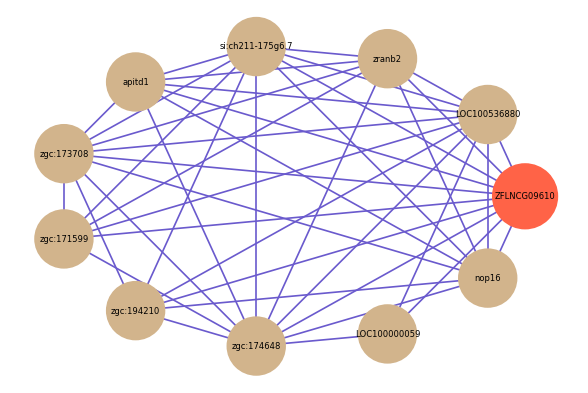
| gene | correlation coefficent |
|---|---|
| LOC100536880 | 0.58 |
| zranb2 | 0.57 |
| si:ch211-175g6.7 | 0.57 |
| apitd1 | 0.56 |
| zgc:173708 | 0.56 |
| zgc:171599 | 0.55 |
| zgc:194210 | 0.55 |
| zgc:174648 | 0.54 |
| LOC100000059 | 0.53 |
| nop16 | 0.52 |
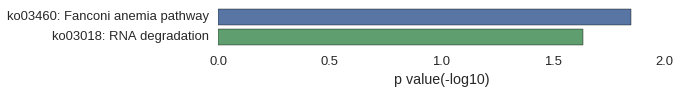
Gene Ontology
Download
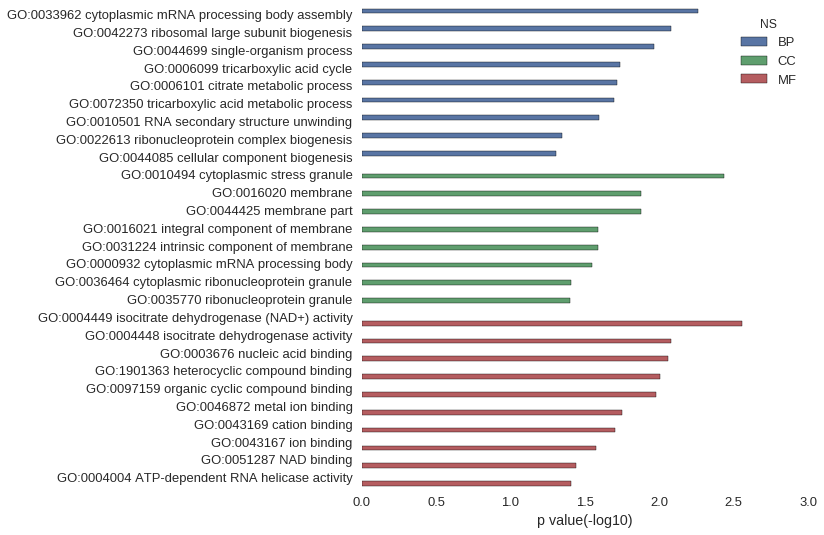
| GO | P value |
|---|---|
| GO:0033962 | 5.53e-03 |
| GO:0042273 | 8.29e-03 |
| GO:0044699 | 1.08e-02 |
| GO:0006099 | 1.83e-02 |
| GO:0006101 | 1.92e-02 |
| GO:0072350 | 2.01e-02 |
| GO:0010501 | 2.56e-02 |
| GO:0022613 | 4.52e-02 |
| GO:0044085 | 4.97e-02 |
| GO:0010494 | 3.69e-03 |
| GO:0016020 | 1.32e-02 |
| GO:0044425 | 1.32e-02 |
| GO:0016021 | 2.57e-02 |
| GO:0031224 | 2.57e-02 |
| GO:0000932 | 2.83e-02 |
| GO:0036464 | 3.90e-02 |
| GO:0035770 | 3.99e-02 |
| GO:0004449 | 2.77e-03 |
| GO:0004448 | 8.29e-03 |
| GO:0003676 | 8.71e-03 |
| GO:1901363 | 9.92e-03 |
| GO:0097159 | 1.06e-02 |
| GO:0046872 | 1.79e-02 |
| GO:0043169 | 1.97e-02 |
| GO:0043167 | 2.67e-02 |
| GO:0051287 | 3.63e-02 |
| GO:0004004 | 3.90e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09610
TTTTAATTAAAAATGCCATCATGGGAGCCAATAGTCAGTTTTAGCAACTGAATCATAATATGCGTTTATT
AATATTAATTATGGTGTCTTTCTTTAAGGAACGATGGACTTATTAGAAATTCCATATTCTGTTTCCTTAT
GTACTGCTTGATGAACAATATTCATCAGAAATGTGGATTAGTTTTAACACCATTTTACTAATTCTAAAAT
TTAAATATCTAAATTGCCATTAAATACCAATGAGATGAAAATCAATGTATTGCATGATGTAAATACAATG
GGTGGATGGATAGAGGTAAAGTTGGTTTTATATTCGGATATTCAGACATTCACCTAAATAATATCTTTCA
TAAAAAGTATTCTTTGGAAATTCTAAATAGGTAAATCATATTAGCAGCAAATGACTATCAAAGTACAACA
TTGTTCACAGTCAAATAAATAGTGTAAATGTTGTTTTCAAATCATACTTCCATGCTAATTCCGATGGAAT
ATTCAAAGTAACATAGTAAACAATATATAGACTGCTAAATGAAAGAATGTGTGAATATAAAACCAACTTT
ACCTCTATGGTGGATGGGGGAGGATGAGCCATCCCTTTAAAATGACATTCACAATGCACTCATGTCTATG
TATATTCTTGGGAATGCCTACTCAGTCTTAAACAGTAGATTTTGTTATTCGTGCTGATGTCTTTAAACCC
ATAAACAATACACACGTGAACCTATCAAATGTAGACTTATTAACTTACTCGTGAACTGCTAAAAGATAAT
TCTGACCTTGACATTAAATGGCAAAGACATCAAATCAACAGCATATTAAACAGATTAACAAGCAAAGTTT
AAAGATAGCATGATCTAACCTCCGTTTTAAATATCTAATAAGTCGCATTAGCATTGTAACACGTTTAAAT
GAATCCTAGAGTACATCATACACAACTATGCAGGTAAAAAGCGAGTAAGCGAGTAAAACTGTTCAGGCGA
TACACAACCAATGTACTCCTAAGAACAAATATCTAAAGATGAGCATCACTAGTTTATATAATATTACTGA
GAAGCACAATAAATTACTTACATAATTTCGTCTTCAAATCCACGGCCAGCATGCCGCAAGCAGACGGATT
CTGTGAATGTCAGACT
TTTTAATTAAAAATGCCATCATGGGAGCCAATAGTCAGTTTTAGCAACTGAATCATAATATGCGTTTATT
AATATTAATTATGGTGTCTTTCTTTAAGGAACGATGGACTTATTAGAAATTCCATATTCTGTTTCCTTAT
GTACTGCTTGATGAACAATATTCATCAGAAATGTGGATTAGTTTTAACACCATTTTACTAATTCTAAAAT
TTAAATATCTAAATTGCCATTAAATACCAATGAGATGAAAATCAATGTATTGCATGATGTAAATACAATG
GGTGGATGGATAGAGGTAAAGTTGGTTTTATATTCGGATATTCAGACATTCACCTAAATAATATCTTTCA
TAAAAAGTATTCTTTGGAAATTCTAAATAGGTAAATCATATTAGCAGCAAATGACTATCAAAGTACAACA
TTGTTCACAGTCAAATAAATAGTGTAAATGTTGTTTTCAAATCATACTTCCATGCTAATTCCGATGGAAT
ATTCAAAGTAACATAGTAAACAATATATAGACTGCTAAATGAAAGAATGTGTGAATATAAAACCAACTTT
ACCTCTATGGTGGATGGGGGAGGATGAGCCATCCCTTTAAAATGACATTCACAATGCACTCATGTCTATG
TATATTCTTGGGAATGCCTACTCAGTCTTAAACAGTAGATTTTGTTATTCGTGCTGATGTCTTTAAACCC
ATAAACAATACACACGTGAACCTATCAAATGTAGACTTATTAACTTACTCGTGAACTGCTAAAAGATAAT
TCTGACCTTGACATTAAATGGCAAAGACATCAAATCAACAGCATATTAAACAGATTAACAAGCAAAGTTT
AAAGATAGCATGATCTAACCTCCGTTTTAAATATCTAATAAGTCGCATTAGCATTGTAACACGTTTAAAT
GAATCCTAGAGTACATCATACACAACTATGCAGGTAAAAAGCGAGTAAGCGAGTAAAACTGTTCAGGCGA
TACACAACCAATGTACTCCTAAGAACAAATATCTAAAGATGAGCATCACTAGTTTATATAATATTACTGA
GAAGCACAATAAATTACTTACATAATTTCGTCTTCAAATCCACGGCCAGCATGCCGCAAGCAGACGGATT
CTGTGAATGTCAGACT