ZFLNCG09724
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR800037 | egg | normal | 41.49 |
| SRR800043 | sphere stage | normal | 32.69 |
| SRR800049 | sphere stage | control treatment | 30.05 |
| SRR1021215 | sphere stage | Mzeomesa | 15.54 |
| SRR1205157 | 5dpf | transgenic sqET20 and neomycin treated 1h and GFP+ | 15.19 |
| SRR1021213 | sphere stage | normal | 15.01 |
| SRR800046 | sphere stage | 5azaCyD treatment | 14.81 |
| SRR1562533 | testis | normal | 12.09 |
| SRR1035984 | 13 hpf | normal | 11.31 |
| SRR527835 | tail | normal | 11.01 |
Express in tissues
Correlated coding gene
Download
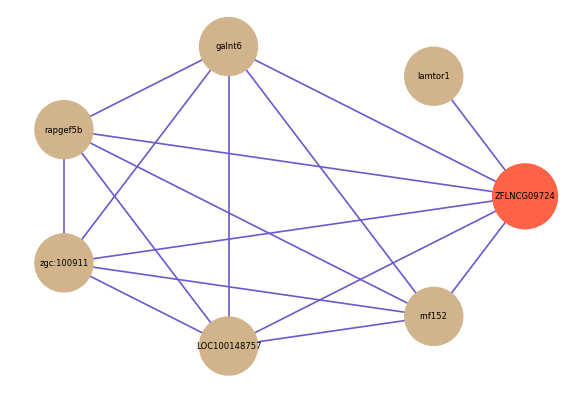
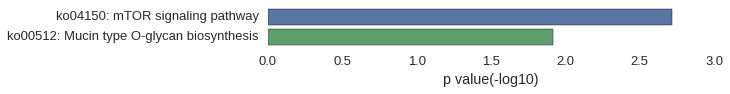
| gene | correlation coefficent |
|---|---|
| lamtor1 | 0.52 |
| galnt6 | 0.52 |
| rapgef5b | 0.52 |
| zgc:100911 | 0.51 |
| LOC100148757 | 0.50 |
| rnf152 | 0.50 |
Gene Ontology
Download
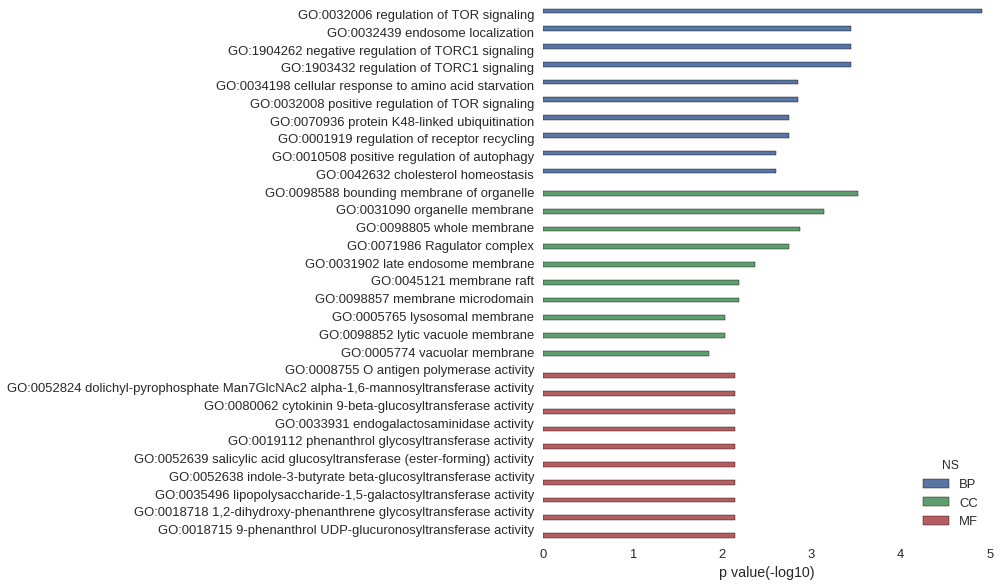
| GO | P value |
|---|---|
| GO:0032006 | 1.21e-05 |
| GO:0032439 | 3.55e-04 |
| GO:1904262 | 3.55e-04 |
| GO:1903432 | 3.55e-04 |
| GO:0034198 | 1.42e-03 |
| GO:0032008 | 1.42e-03 |
| GO:0070936 | 1.78e-03 |
| GO:0001919 | 1.78e-03 |
| GO:0010508 | 2.49e-03 |
| GO:0042632 | 2.49e-03 |
| GO:0098588 | 2.96e-04 |
| GO:0031090 | 7.27e-04 |
| GO:0098805 | 1.32e-03 |
| GO:0071986 | 1.78e-03 |
| GO:0031902 | 4.26e-03 |
| GO:0045121 | 6.38e-03 |
| GO:0098857 | 6.38e-03 |
| GO:0005765 | 9.21e-03 |
| GO:0098852 | 9.21e-03 |
| GO:0005774 | 1.41e-02 |
| GO:0008755 | 7.09e-03 |
| GO:0052824 | 7.09e-03 |
| GO:0080062 | 7.09e-03 |
| GO:0033931 | 7.09e-03 |
| GO:0019112 | 7.09e-03 |
| GO:0052639 | 7.09e-03 |
| GO:0052638 | 7.09e-03 |
| GO:0035496 | 7.09e-03 |
| GO:0018718 | 7.09e-03 |
| GO:0018715 | 7.09e-03 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09724
GCGTGTCCTCAAATCTGCCTCTGAAACACAATGGGATCACAATCACTGCCTCCTTTGGGACAGCAGAGAA
CAAACTACACAGACGACAACAACAGAATTTAGGTCATTACAGAGGCATATCGACTCCTGCTGAATCAGCT
GCAGCTTAAAAGAACAAATAGATGTAGACAGAAGAGTAACGGGCCCTGAGGTTTGACTGCATTACGTTGT
TGTGTGTAATGGGAGAGTGTGTGTTGGACCGCTATGAAGAAGATGGGTTTCAGAGTTGTGAAGGGTTTGA
CAACTGGGCCTCATTTAGC
GCGTGTCCTCAAATCTGCCTCTGAAACACAATGGGATCACAATCACTGCCTCCTTTGGGACAGCAGAGAA
CAAACTACACAGACGACAACAACAGAATTTAGGTCATTACAGAGGCATATCGACTCCTGCTGAATCAGCT
GCAGCTTAAAAGAACAAATAGATGTAGACAGAAGAGTAACGGGCCCTGAGGTTTGACTGCATTACGTTGT
TGTGTGTAATGGGAGAGTGTGTGTTGGACCGCTATGAAGAAGATGGGTTTCAGAGTTGTGAAGGGTTTGA
CAACTGGGCCTCATTTAGC