ZFLNCG09726
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR658537 | bud | Gata6 morphant | 29.21 |
| SRR658547 | 6 somite | Gata5/6 morphant | 27.00 |
| SRR658543 | 6 somite | Gata5 morphan | 26.28 |
| SRR658541 | 6 somite | normal | 23.37 |
| SRR658545 | 6 somite | Gata6 morphant | 22.39 |
| SRR658539 | bud | Gata5/6 morphant | 22.24 |
| SRR658535 | bud | Gata5 morphant | 20.80 |
| SRR038627 | embryo | control morpholino | 20.49 |
| SRR516132 | skin | male and 5 month | 19.35 |
| SRR749514 | 24 hpf | normal | 18.96 |
Express in tissues
Correlated coding gene
Download
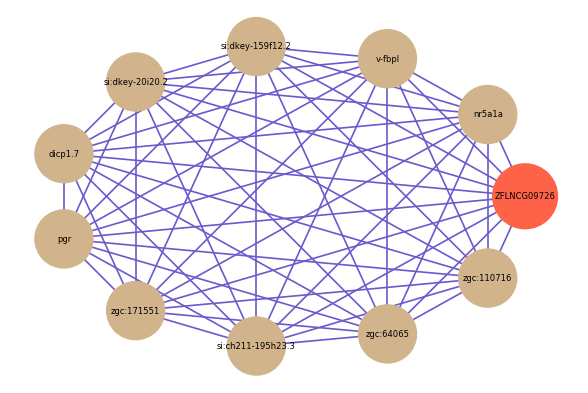
| gene | correlation coefficent |
|---|---|
| nr5a1a | 0.72 |
| v-fbpl | 0.71 |
| si:dkey-159f12.2 | 0.69 |
| si:dkey-20i20.2 | 0.67 |
| dicp1.7 | 0.67 |
| pgr | 0.67 |
| zgc:171551 | 0.66 |
| si:ch211-195h23.3 | 0.65 |
| zgc:64065 | 0.65 |
| zgc:110716 | 0.65 |
Gene Ontology
Download
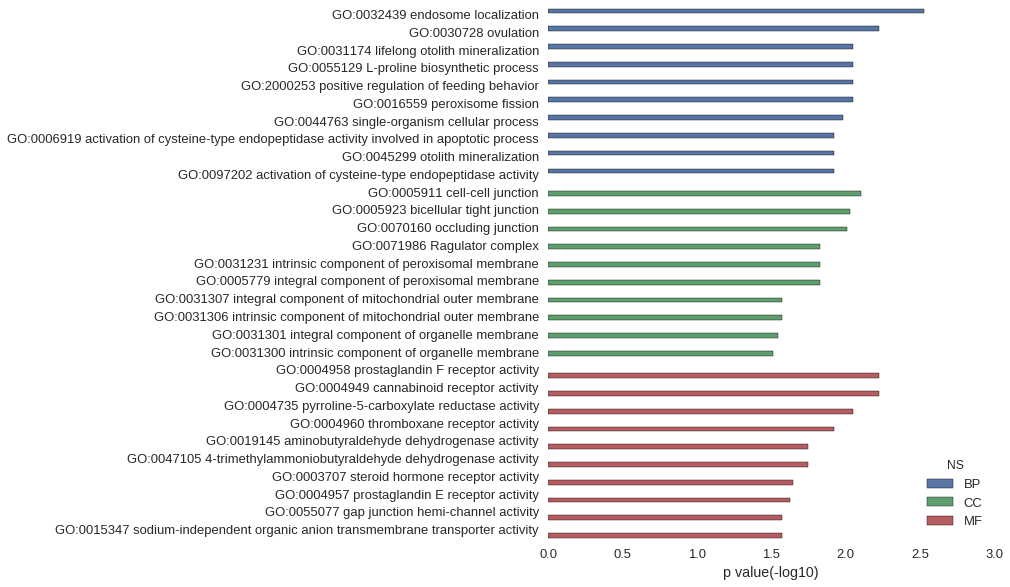
| GO | P value |
|---|---|
| GO:0032439 | 2.98e-03 |
| GO:0030728 | 5.96e-03 |
| GO:0031174 | 8.93e-03 |
| GO:0055129 | 8.93e-03 |
| GO:2000253 | 8.93e-03 |
| GO:0016559 | 8.93e-03 |
| GO:0044763 | 1.04e-02 |
| GO:0006919 | 1.19e-02 |
| GO:0045299 | 1.19e-02 |
| GO:0097202 | 1.19e-02 |
| GO:0005911 | 7.85e-03 |
| GO:0005923 | 9.36e-03 |
| GO:0070160 | 9.73e-03 |
| GO:0071986 | 1.48e-02 |
| GO:0031231 | 1.48e-02 |
| GO:0005779 | 1.48e-02 |
| GO:0031307 | 2.66e-02 |
| GO:0031306 | 2.66e-02 |
| GO:0031301 | 2.83e-02 |
| GO:0031300 | 3.07e-02 |
| GO:0004958 | 5.96e-03 |
| GO:0004949 | 5.96e-03 |
| GO:0004735 | 8.93e-03 |
| GO:0004960 | 1.19e-02 |
| GO:0019145 | 1.78e-02 |
| GO:0047105 | 1.78e-02 |
| GO:0003707 | 2.26e-02 |
| GO:0004957 | 2.36e-02 |
| GO:0055077 | 2.66e-02 |
| GO:0015347 | 2.66e-02 |
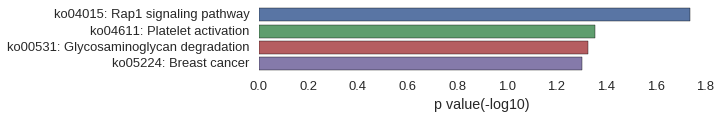
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09726
ATGACGAGCGAAAACACCCGACTCTGTGTCTGCTCTGTGGGGTGATGTTGTGTGCTCAGAGCTCCTGCTG
TCATGAGCAGCTCAACGGCGAGGAAGTGGGCGCCTGTACGGGTCACGCTGCCATCTGTGGGGCTGGAGTG
GGACTGTTTCTCAGGTAAGAGAAACCAAAAAAACATTTTCTCCTTATTTGAGTGACAAGATAAAGGAATC
TTTAAAAGATCACAAATGTGTTGCATATGTTTATTAAACCTTTGACAATAACAGCTCTACATGTATGGCT
TGTTTTTACTGAGTAAAAAAGCAGGATGTCTAGTCTCGTCTTTACATTGACTTAACATGTAAACCACTTG
CACTTGATGCTTAATCCGCGTCTAGTGTGAATGCACCATTACATAAATGAATCCTTTTCACAAGACTGTT
ATGATGTGTTTAACGGTCATCAGAATCCTAAATGCTTTTTTTAATATATTATTTAAAAAAAAAAAACAGA
TAAACATTTATATATGTATTATATCAAACTAATTACCACTTATCTAAGGTGTTTCTAAAGCAACATGTAT
GGGGATGAGAGTAAATTTGACGTTATTCTCTCCTCTGGCTGTTGTTATCAGTCTGGCACTATTATTTTTT
TCTTTTTCTGAAACTCTTGATAACGCCATTGAGGACTTTTTCTTTTATTATTTCAGCAAGTAGGGTTGTA
AAAAATTTAACAACACTCTAAGGTTACCCAACTGTGACTGTCACTTAACAACTTAAAAGAAAAGGGTTGG
TGGATTTTTATAAAATACAAATAAATAGGGGAAAAAGTTTGAAAAAGCTGAGTGACAGTATATTGTTAGG
AAATTTTTGACATCCCTACACTTCATCGAAAGAACTTTAATGACAGAACAGTGCAGCGAGGTTGTGCTTC
TCATGAACTTTCGTTTTCCAAATGCCAAAACACACTCAAAAAGAACTTCATTTGGGCAGATTTTGTTAGA
AATTATGCTATTTCAGTTATTATTTCTGATTTCATTTGAAGTTTCCTAAGGTCTTAAATGTGTTGTTTGT
GTATTTCTAAACAGGGTTCGAGAGTGTGAGATTGTCTTGATGGCCAGTAAGACACGTGGCAGTCCCTATC
CAGCACCCTACTTAGACGACTATGGAGAAACAGACCCT
ATGACGAGCGAAAACACCCGACTCTGTGTCTGCTCTGTGGGGTGATGTTGTGTGCTCAGAGCTCCTGCTG
TCATGAGCAGCTCAACGGCGAGGAAGTGGGCGCCTGTACGGGTCACGCTGCCATCTGTGGGGCTGGAGTG
GGACTGTTTCTCAGGTAAGAGAAACCAAAAAAACATTTTCTCCTTATTTGAGTGACAAGATAAAGGAATC
TTTAAAAGATCACAAATGTGTTGCATATGTTTATTAAACCTTTGACAATAACAGCTCTACATGTATGGCT
TGTTTTTACTGAGTAAAAAAGCAGGATGTCTAGTCTCGTCTTTACATTGACTTAACATGTAAACCACTTG
CACTTGATGCTTAATCCGCGTCTAGTGTGAATGCACCATTACATAAATGAATCCTTTTCACAAGACTGTT
ATGATGTGTTTAACGGTCATCAGAATCCTAAATGCTTTTTTTAATATATTATTTAAAAAAAAAAAACAGA
TAAACATTTATATATGTATTATATCAAACTAATTACCACTTATCTAAGGTGTTTCTAAAGCAACATGTAT
GGGGATGAGAGTAAATTTGACGTTATTCTCTCCTCTGGCTGTTGTTATCAGTCTGGCACTATTATTTTTT
TCTTTTTCTGAAACTCTTGATAACGCCATTGAGGACTTTTTCTTTTATTATTTCAGCAAGTAGGGTTGTA
AAAAATTTAACAACACTCTAAGGTTACCCAACTGTGACTGTCACTTAACAACTTAAAAGAAAAGGGTTGG
TGGATTTTTATAAAATACAAATAAATAGGGGAAAAAGTTTGAAAAAGCTGAGTGACAGTATATTGTTAGG
AAATTTTTGACATCCCTACACTTCATCGAAAGAACTTTAATGACAGAACAGTGCAGCGAGGTTGTGCTTC
TCATGAACTTTCGTTTTCCAAATGCCAAAACACACTCAAAAAGAACTTCATTTGGGCAGATTTTGTTAGA
AATTATGCTATTTCAGTTATTATTTCTGATTTCATTTGAAGTTTCCTAAGGTCTTAAATGTGTTGTTTGT
GTATTTCTAAACAGGGTTCGAGAGTGTGAGATTGTCTTGATGGCCAGTAAGACACGTGGCAGTCCCTATC
CAGCACCCTACTTAGACGACTATGGAGAAACAGACCCT