ZFLNCG09881
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023146 | head kidney | normal | 20.23 |
| ERR023143 | swim bladder | normal | 12.38 |
| SRR1205160 | 5dpf | transgenic sqET20 and GFP+ | 9.67 |
| SRR519752 | 7 dpf | VD3 treatment | 8.73 |
| SRR519747 | 6 dpf | VD3 treatment | 7.59 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 7.35 |
| SRR519737 | 2 dpf | VD3 treatment | 7.27 |
| SRR941753 | posterior pectoral fin | normal | 7.13 |
| SRR1205169 | 5dpf | transgenic sqET20 and neomycin treated 5h and GFP+ | 6.99 |
| SRR519732 | 7 dpf | FETOH treatment | 6.29 |
Express in tissues
Correlated coding gene
Download
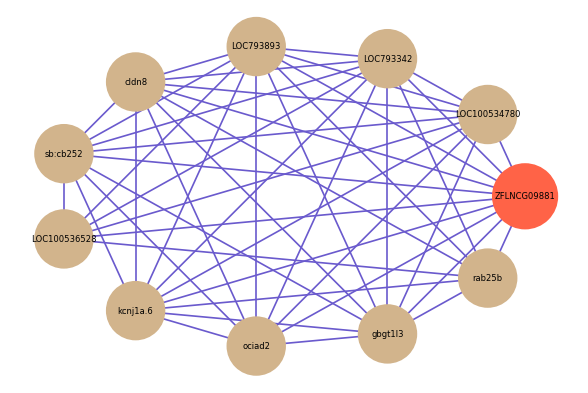
| gene | correlation coefficent |
|---|---|
| LOC100534780 | 0.68 |
| LOC793342 | 0.67 |
| LOC793893 | 0.58 |
| cldn8 | 0.57 |
| sb:cb252 | 0.57 |
| LOC100536528 | 0.56 |
| kcnj1a.6 | 0.56 |
| rab25b | 0.55 |
| ociad2 | 0.55 |
| gbgt1l3 | 0.55 |
Gene Ontology
Download

| GO | P value |
|---|---|
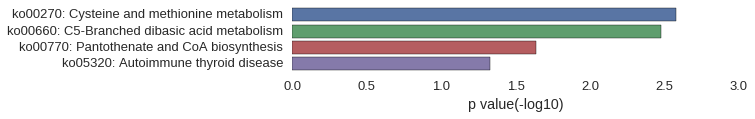
| GO:0044260 | 1.28e-03 |
| GO:0010107 | 1.37e-03 |
| GO:0046381 | 2.20e-03 |
| GO:0006054 | 6.59e-03 |
| GO:0033353 | 6.59e-03 |
| GO:0044237 | 7.43e-03 |
| GO:0048730 | 8.78e-03 |
| GO:0046498 | 8.78e-03 |
| GO:0071805 | 9.50e-03 |
| GO:0071804 | 9.50e-03 |
| GO:0031012 | 4.96e-03 |
| GO:0044422 | 1.04e-02 |
| GO:0044446 | 1.78e-02 |
| GO:0005615 | 2.92e-02 |
| GO:0005911 | 3.64e-02 |
| GO:0005578 | 3.64e-02 |
| GO:0016021 | 4.35e-02 |
| GO:0031224 | 4.44e-02 |
| GO:0015272 | 1.67e-04 |
| GO:0005242 | 7.03e-04 |
| GO:0030338 | 2.20e-03 |
| GO:0047613 | 4.40e-03 |
| GO:0015165 | 4.40e-03 |
| GO:0004145 | 4.40e-03 |
| GO:0005338 | 6.59e-03 |
| GO:0008453 | 6.59e-03 |
| GO:0016716 | 8.78e-03 |
| GO:0016802 | 8.78e-03 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09881
TGTCACATGGCACTATATGGTCTTATAATCCTGCAGCCCATCATAAGTGTCTCCTGACCTTCTCCCTCTG
CGCCTCTGAGCTCTCCAGCTCCCCCTGCAGGCGGGCGATGGTGCCGCTTAACTCCTGACGCTGCTCTGCT
GCCTCACGACGGTCCTGCTTCAGGATGTCCATCAGCGCCTACACACACACAACATCAGACACACATGAAA
TCAGACACACACAACATCACACACACATCAGACACACACACACACACACACACACGAAATCAGAGACACA
CACGACATCAGATACGCACAACATCACACACAACATCAGACACACACACACACACGAAATCAGAGACACA
CACGACATCAGATACACACACAACATCACACACACAACATCAGAAACACACACACACACACGAAATCAGA
GACACACACAACATCAGATACACACACAACATCACACACACAACATCAGACACACACACACACACACAAA
ATCAGAGACACACACGACATCAGATACACACACAACATCACACACACAACCTCAGACACACACACACACA
CACGAAATCAGAGACACACACGACATCAGATACACACACAACATCACACACACAACATC
TGTCACATGGCACTATATGGTCTTATAATCCTGCAGCCCATCATAAGTGTCTCCTGACCTTCTCCCTCTG
CGCCTCTGAGCTCTCCAGCTCCCCCTGCAGGCGGGCGATGGTGCCGCTTAACTCCTGACGCTGCTCTGCT
GCCTCACGACGGTCCTGCTTCAGGATGTCCATCAGCGCCTACACACACACAACATCAGACACACATGAAA
TCAGACACACACAACATCACACACACATCAGACACACACACACACACACACACACGAAATCAGAGACACA
CACGACATCAGATACGCACAACATCACACACAACATCAGACACACACACACACACGAAATCAGAGACACA
CACGACATCAGATACACACACAACATCACACACACAACATCAGAAACACACACACACACACGAAATCAGA
GACACACACAACATCAGATACACACACAACATCACACACACAACATCAGACACACACACACACACACAAA
ATCAGAGACACACACGACATCAGATACACACACAACATCACACACACAACCTCAGACACACACACACACA
CACGAAATCAGAGACACACACGACATCAGATACACACACAACATCACACACACAACATC