ZFLNCG09945
Basic Information
Chromesome: chr18
Start: 7034441
End: 7035618
Transcript: ZFLNCT15313 ZFLNCT15314
Known as: ENSDARG00000094719 LOC101884954
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| ERR023146 | head kidney | normal | 544.75 |
| ERR023143 | swim bladder | normal | 514.73 |
| SRR1562529 | intestine and pancreas | normal | 512.49 |
| ERR023145 | heart | normal | 376.97 |
| SRR1299124 | caudal fin | Zero day time after treatment | 273.18 |
| SRR1647683 | spleen | SVCV treatment | 244.90 |
| SRR1647680 | head kidney | SVCV treatment | 207.67 |
| SRR516124 | skin | male and 3.5 year | 180.81 |
| SRR1562528 | eye | normal | 173.28 |
| SRR516125 | skin | male and 3.5 year | 169.50 |
Express in tissues
Correlated coding gene
Download
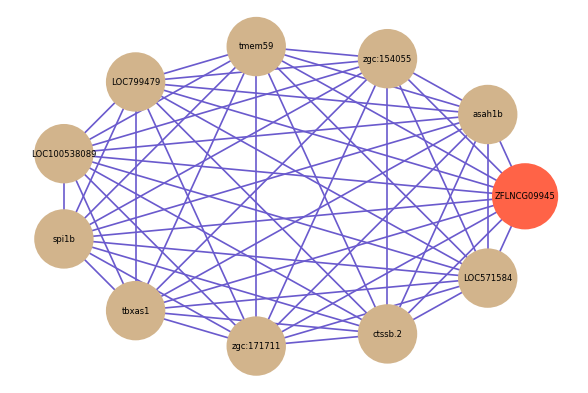
| gene | correlation coefficent |
|---|---|
| asah1b | 0.65 |
| zgc:154055 | 0.65 |
| tmem59 | 0.65 |
| LOC799479 | 0.63 |
| LOC100538089 | 0.63 |
| spi1b | 0.63 |
| tbxas1 | 0.63 |
| zgc:171711 | 0.62 |
| ctssb.2 | 0.62 |
| LOC571584 | 0.62 |
Gene Ontology
Download
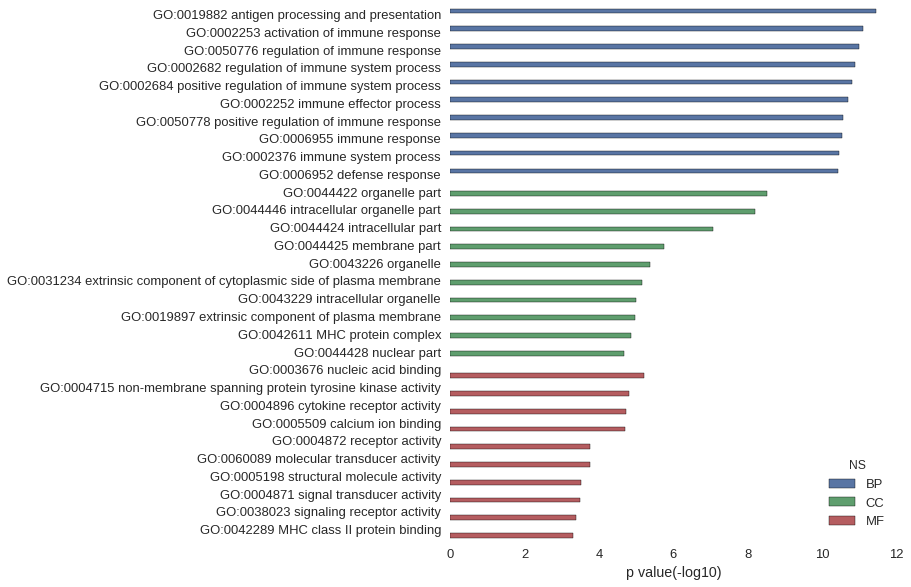
| GO | P value |
|---|---|
| GO:0019882 | 3.52e-12 |
| GO:0002253 | 8.02e-12 |
| GO:0050776 | 1.01e-11 |
| GO:0002682 | 1.31e-11 |
| GO:0002684 | 1.61e-11 |
| GO:0002252 | 2.05e-11 |
| GO:0050778 | 2.75e-11 |
| GO:0006955 | 2.95e-11 |
| GO:0002376 | 3.64e-11 |
| GO:0006952 | 3.82e-11 |
| GO:0044422 | 3.13e-09 |
| GO:0044446 | 6.46e-09 |
| GO:0044424 | 8.86e-08 |
| GO:0044425 | 1.80e-06 |
| GO:0043226 | 4.38e-06 |
| GO:0031234 | 6.97e-06 |
| GO:0043229 | 9.91e-06 |
| GO:0019897 | 1.06e-05 |
| GO:0042611 | 1.38e-05 |
| GO:0044428 | 2.08e-05 |
| GO:0003676 | 5.99e-06 |
| GO:0004715 | 1.55e-05 |
| GO:0004896 | 1.87e-05 |
| GO:0005509 | 1.95e-05 |
| GO:0004872 | 1.70e-04 |
| GO:0060089 | 1.70e-04 |
| GO:0005198 | 2.99e-04 |
| GO:0004871 | 3.28e-04 |
| GO:0038023 | 4.15e-04 |
| GO:0042289 | 5.03e-04 |
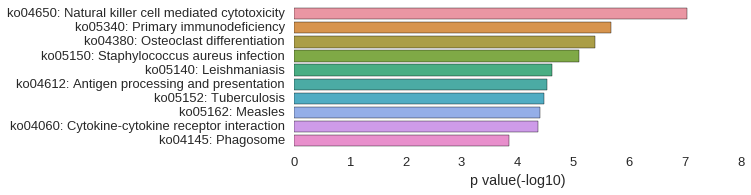
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG09945
CATCATGAAGGACGTGGTGGTCATGTAAATCCTTGCTGCCAGGGGCGAGGACATCTGGCCCAGCGCCATT
CAAAGGGGAACCTCAGCATTTGCATGAGTTCCTCTTTGTCTTGCGCTGGTGTCCAGATGCGCAGAGGCCT
GACTGCTGCGAAGATGATGTTTAAGTTGAACCTTCAAGTCTTGGAAGATACGAAGAGTCAAGTCTACTAT
CATGATCACATCTAAATAAGCAGAAGTCAGACCGATGTGAAGCCTGATGATTTTTGTTGAATTCTTAAGT
CTATAAACCTACGAAGTCTCAATTCTACTATTATCACATCTATATACGCAGAGTGAAGTCTACGAAGTCT
CAAGTCTACTAATATGAACGCATCTATATACGCAGAGTCTATCAAGTCTACAAAGTCTCAAGTCTACTAT
TATGATCGCATCTATATACACAGAGCCCAGAACTGGTGTGAAGCCTGACGTTTTTGCCGAATCCTCAAGT
CTATGAAGCCTACTGTTATGACCACGTCTCGATACGCAGAGCCCAGAACTGGTGTGAAGCCTGATGTTTT
TCCGTCGAACCCTCCTCTATGAAGTCTCAAATCTACTATTATGATCGCATCTATACAAGCAGAACCCAGA
ACTGGTGTGAAGCCTGACGTTTTTTGCCGAACCCTCAAGTCTATAAAGCCTACTGTTTATGACCACGTCT
CGATACGCAAAGCCCAGAACTGGTATGAAGTCTGAGTTGTTTTTTTTTGTCGAACCCTTGTATGTGAAAT
TTTAAATCTACTATTATGGATGCATTTATACAATTAGAGTCCTAACTAGTGTGAAGCCGGATTATTTTTG
TCGGACCCTCAAGTCAACGAAGCAGTAAAGTCTCAAATTGATTACCATGGTTAAATTCAGATAGAGAGAA
GCCTATTGTATGTGCAGTGTCAAAAATTGAAACGTTTTGGCTTCTGCAATACTTGGTTTCAGCTGGCTTG
GGGCCCAGCTGAGGTAAAAACCCTGTCCCCTTCACCCAAACCTACCAAAAAGACAACAAAAAAAATGAAT
AAACTCTGCAACACAATTACAAATGGTACTGTTTGTTTATTTATTACGGGTGTATTATTGGTAACATACA
AATTCAGAGTTATTGTATGACTCATGGAGAACTAAACCATTTATTAGTGAATTGCTG
CATCATGAAGGACGTGGTGGTCATGTAAATCCTTGCTGCCAGGGGCGAGGACATCTGGCCCAGCGCCATT
CAAAGGGGAACCTCAGCATTTGCATGAGTTCCTCTTTGTCTTGCGCTGGTGTCCAGATGCGCAGAGGCCT
GACTGCTGCGAAGATGATGTTTAAGTTGAACCTTCAAGTCTTGGAAGATACGAAGAGTCAAGTCTACTAT
CATGATCACATCTAAATAAGCAGAAGTCAGACCGATGTGAAGCCTGATGATTTTTGTTGAATTCTTAAGT
CTATAAACCTACGAAGTCTCAATTCTACTATTATCACATCTATATACGCAGAGTGAAGTCTACGAAGTCT
CAAGTCTACTAATATGAACGCATCTATATACGCAGAGTCTATCAAGTCTACAAAGTCTCAAGTCTACTAT
TATGATCGCATCTATATACACAGAGCCCAGAACTGGTGTGAAGCCTGACGTTTTTGCCGAATCCTCAAGT
CTATGAAGCCTACTGTTATGACCACGTCTCGATACGCAGAGCCCAGAACTGGTGTGAAGCCTGATGTTTT
TCCGTCGAACCCTCCTCTATGAAGTCTCAAATCTACTATTATGATCGCATCTATACAAGCAGAACCCAGA
ACTGGTGTGAAGCCTGACGTTTTTTGCCGAACCCTCAAGTCTATAAAGCCTACTGTTTATGACCACGTCT
CGATACGCAAAGCCCAGAACTGGTATGAAGTCTGAGTTGTTTTTTTTTGTCGAACCCTTGTATGTGAAAT
TTTAAATCTACTATTATGGATGCATTTATACAATTAGAGTCCTAACTAGTGTGAAGCCGGATTATTTTTG
TCGGACCCTCAAGTCAACGAAGCAGTAAAGTCTCAAATTGATTACCATGGTTAAATTCAGATAGAGAGAA
GCCTATTGTATGTGCAGTGTCAAAAATTGAAACGTTTTGGCTTCTGCAATACTTGGTTTCAGCTGGCTTG
GGGCCCAGCTGAGGTAAAAACCCTGTCCCCTTCACCCAAACCTACCAAAAAGACAACAAAAAAAATGAAT
AAACTCTGCAACACAATTACAAATGGTACTGTTTGTTTATTTATTACGGGTGTATTATTGGTAACATACA
AATTCAGAGTTATTGTATGACTCATGGAGAACTAAACCATTTATTAGTGAATTGCTG