ZFLNCG10185
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR372787 | 2-4 cell | normal | 0.88 |
| SRR1049814 | 256 cell | normal | 0.83 |
| SRR1621206 | embryo | normal | 0.56 |
| SRR372789 | 1K cell | normal | 0.32 |
| SRR748489 | egg | normal | 0.28 |
| SRR372791 | dome | normal | 0.28 |
| SRR748490 | 1 kcell | normal | 0.27 |
| SRR748491 | germ ring | normal | 0.22 |
| SRR800046 | sphere stage | 5azaCyD treatment | 0.20 |
| SRR1021215 | sphere stage | Mzeomesa | 0.15 |
Express in tissues
Correlated coding gene
Download
| gene | correlation coefficent |
|---|---|
| LOC100331830 | 0.72 |
| LOC100537183 | 0.62 |
| si:dkey-179j5.5 | 0.57 |
| LOC100331909 | 0.55 |
| si:ch211-190p8.2 | 0.54 |
| LOC101882162 | 0.53 |
| LOC100535847 | 0.53 |
| LOC101883986 | 0.52 |
| si:ch211-250e5.9 | 0.52 |
| LOC100537146 | 0.52 |
Gene Ontology
Download
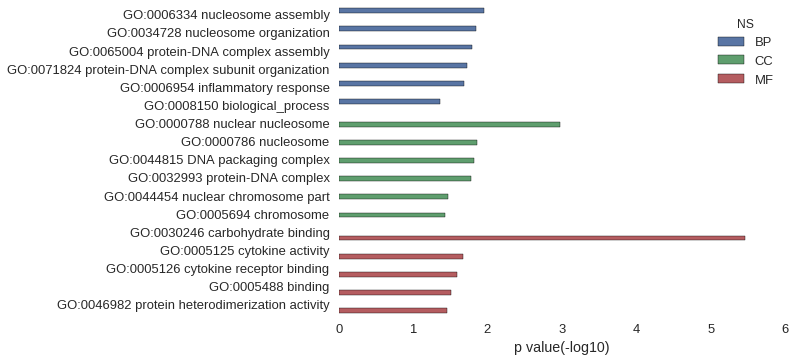
| GO | P value |
|---|---|
| GO:0006334 | 1.13e-02 |
| GO:0034728 | 1.41e-02 |
| GO:0065004 | 1.59e-02 |
| GO:0071824 | 1.87e-02 |
| GO:0006954 | 2.04e-02 |
| GO:0008150 | 4.40e-02 |
| GO:0000788 | 1.07e-03 |
| GO:0000786 | 1.38e-02 |
| GO:0044815 | 1.52e-02 |
| GO:0032993 | 1.69e-02 |
| GO:0044454 | 3.43e-02 |
| GO:0005694 | 3.68e-02 |
| GO:0030246 | 3.45e-06 |
| GO:0005125 | 2.11e-02 |
| GO:0005126 | 2.60e-02 |
| GO:0005488 | 3.07e-02 |
| GO:0046982 | 3.47e-02 |
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10185
GCCATGCTGCTGACAAATTTAGCTCTTGTCCATGTGTCATGTTTGGAAAAACCAAGTCAAGTCTAACTTT
GTTAAGCTAAGTTCAATTTGACTACAAAATAAAATACAAGACAGCATGACACTTAACCAGACCTATCTCT
ATTTACACATGTAAATGATAATTTTTGGTCAAATATAAACAAACAACTTTTTTTTTTTTTTTTTTTTAAG
TGCAGTTAAGTGAGCATCGTTACTCCCTAATTTTCATGCAGAATTTTGTGACTTTTCAGAGAGTAAATGT
TTAAGTACACTGGAAAAAAAATGTGGTTAATCATGGAGGCGGTCATTTTAAAGACTGTCTCAGTTTCCTG
TCCACTGAATACAGGAACTGCTTGAGATGCACATCTAGATTCTATAGTCACTTTTATTGGCTAATGACCA
ATCAGAAACTGTCTCGGAGAAATGTTATACTATAACACACAGGTGTGTAAAACTCTTCAACATATTAGAA
AAAAAGAGTCTCTGCTGTGAAGTAAACCTAGTTTAAATACTAAGTTTAGTTAATGGCAAGCATTGATTTT
TCACAGTCTTTGTTTCCAGGTGAAATTTACATAGAATCTGGCTTGCATCAAGTTTTAGGCAGATGAGATG
TAATATCATATCCTTCTTCCAAGGATAAGGGATTAAAGGCCTATTTGAAGCAGGAACAGATGACAACAAA
GTCTAATTACAGGTTTACAGTTTTAAGATTTCAAGTTTTTAAGATATTAATGCTCTCTTCCAGAAGGCCC
ATGGGATCTGACTCTTCTTACCTTTCCTTGCTCTTTCAAGTCTACATCCCATCGTGAAGCCAGCAGTTGT
TACATGAGGGTCACCTCAGGGTACCATCCCAGACAGGCACAAAATCACTTTCCTGGACCTTTTCATTCAC
TGTTTCTCAATGGTAGAATGATCTCCTCAACCTCTTGTGAAATGTCAAATCCCTGACAATACTCATGAGT
TTTCACTTTTCTCTTATCTTC
GCCATGCTGCTGACAAATTTAGCTCTTGTCCATGTGTCATGTTTGGAAAAACCAAGTCAAGTCTAACTTT
GTTAAGCTAAGTTCAATTTGACTACAAAATAAAATACAAGACAGCATGACACTTAACCAGACCTATCTCT
ATTTACACATGTAAATGATAATTTTTGGTCAAATATAAACAAACAACTTTTTTTTTTTTTTTTTTTTAAG
TGCAGTTAAGTGAGCATCGTTACTCCCTAATTTTCATGCAGAATTTTGTGACTTTTCAGAGAGTAAATGT
TTAAGTACACTGGAAAAAAAATGTGGTTAATCATGGAGGCGGTCATTTTAAAGACTGTCTCAGTTTCCTG
TCCACTGAATACAGGAACTGCTTGAGATGCACATCTAGATTCTATAGTCACTTTTATTGGCTAATGACCA
ATCAGAAACTGTCTCGGAGAAATGTTATACTATAACACACAGGTGTGTAAAACTCTTCAACATATTAGAA
AAAAAGAGTCTCTGCTGTGAAGTAAACCTAGTTTAAATACTAAGTTTAGTTAATGGCAAGCATTGATTTT
TCACAGTCTTTGTTTCCAGGTGAAATTTACATAGAATCTGGCTTGCATCAAGTTTTAGGCAGATGAGATG
TAATATCATATCCTTCTTCCAAGGATAAGGGATTAAAGGCCTATTTGAAGCAGGAACAGATGACAACAAA
GTCTAATTACAGGTTTACAGTTTTAAGATTTCAAGTTTTTAAGATATTAATGCTCTCTTCCAGAAGGCCC
ATGGGATCTGACTCTTCTTACCTTTCCTTGCTCTTTCAAGTCTACATCCCATCGTGAAGCCAGCAGTTGT
TACATGAGGGTCACCTCAGGGTACCATCCCAGACAGGCACAAAATCACTTTCCTGGACCTTTTCATTCAC
TGTTTCTCAATGGTAGAATGATCTCCTCAACCTCTTGTGAAATGTCAAATCCCTGACAATACTCATGAGT
TTTCACTTTTCTCTTATCTTC

