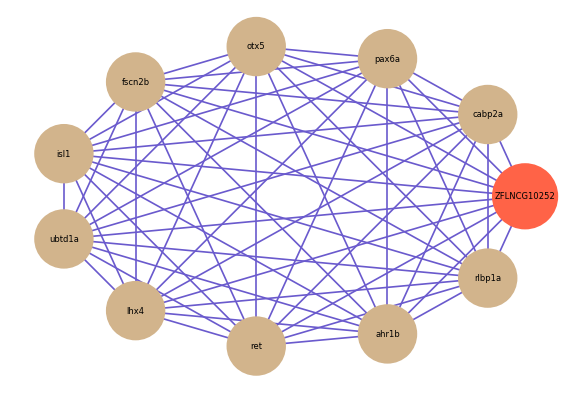
ZFLNCG10252
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1562528 | eye | normal | 276.75 |
| ERR594416 | retina | transgenic flk1 | 170.64 |
| SRR527834 | head | normal | 83.15 |
| SRR1048059 | pineal gland | light | 47.19 |
| SRR1291417 | 5 dpi | injected marinum | 35.19 |
| ERR594417 | retina | normal | 27.41 |
| SRR1648856 | brain | normal | 26.23 |
| SRR592698 | pineal gland | normal | 23.11 |
| SRR726542 | 5 dpf | infection with control | 19.81 |
| SRR1291414 | 5 dpi | normal | 19.34 |
Express in tissues
Correlated coding gene
Download
Gene Ontology
Download
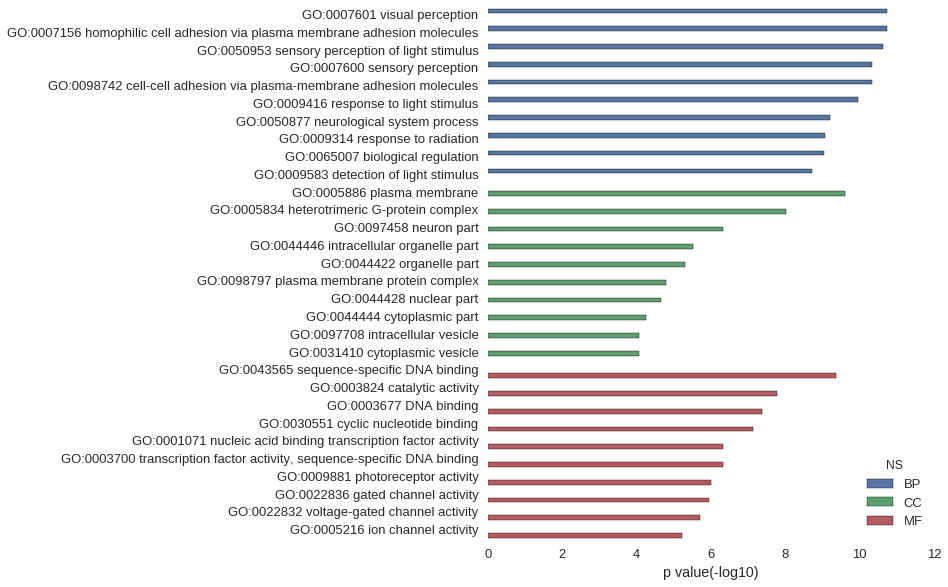
| GO | P value |
|---|---|
| GO:0007601 | 1.83e-11 |
| GO:0007156 | 1.90e-11 |
| GO:0050953 | 2.38e-11 |
| GO:0007600 | 4.64e-11 |
| GO:0098742 | 4.66e-11 |
| GO:0009416 | 1.08e-10 |
| GO:0050877 | 6.11e-10 |
| GO:0009314 | 8.43e-10 |
| GO:0065007 | 9.03e-10 |
| GO:0009583 | 1.87e-09 |
| GO:0005886 | 2.51e-10 |
| GO:0005834 | 9.76e-09 |
| GO:0097458 | 4.82e-07 |
| GO:0044446 | 2.95e-06 |
| GO:0044422 | 5.00e-06 |
| GO:0098797 | 1.60e-05 |
| GO:0044428 | 2.13e-05 |
| GO:0044444 | 5.56e-05 |
| GO:0097708 | 8.42e-05 |
| GO:0031410 | 8.42e-05 |
| GO:0043565 | 4.20e-10 |
| GO:0003824 | 1.62e-08 |
| GO:0003677 | 4.11e-08 |
| GO:0030551 | 7.56e-08 |
| GO:0001071 | 4.62e-07 |
| GO:0003700 | 4.62e-07 |
| GO:0009881 | 9.95e-07 |
| GO:0022836 | 1.15e-06 |
| GO:0022832 | 2.00e-06 |
| GO:0005216 | 5.81e-06 |
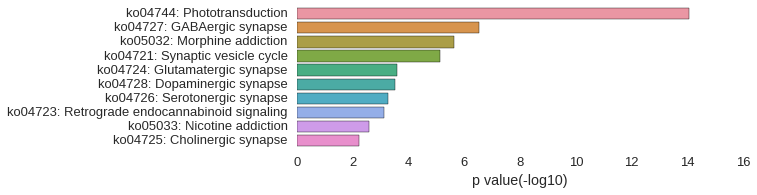
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10252
AATGTACTGCGACTATTTGTGCTGCCGAGAGTTTAAGAACCAAAATCCCAAATCTAGACTGACGAAATAA
AACCTGCTGAATACAAAACATGAGAGTTGATATATGAGTGGTGCTAAAAAACAGTGATCGTATCTGAACT
ATCGCCGACACTGTATCCAGTGATAATCTCCAGGAATGTCATCAATATCAATACAATAAATAGTGAAAGG
TTGTGTTTGGGTGAGAGCGAGAGTGATATTAAGCTCTCGGCGTCAAGTGCAGTTTAGCTGCCGAACAGGA
TTACAACA
AATGTACTGCGACTATTTGTGCTGCCGAGAGTTTAAGAACCAAAATCCCAAATCTAGACTGACGAAATAA
AACCTGCTGAATACAAAACATGAGAGTTGATATATGAGTGGTGCTAAAAAACAGTGATCGTATCTGAACT
ATCGCCGACACTGTATCCAGTGATAATCTCCAGGAATGTCATCAATATCAATACAATAAATAGTGAAAGG
TTGTGTTTGGGTGAGAGCGAGAGTGATATTAAGCTCTCGGCGTCAAGTGCAGTTTAGCTGCCGAACAGGA
TTACAACA