ZFLNCG10278
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR519752 | 7 dpf | VD3 treatment | 241.89 |
| SRR372802 | 5 dpf | normal | 233.02 |
| SRR519732 | 7 dpf | FETOH treatment | 192.63 |
| SRR519727 | 6 dpf | FETOH treatment | 160.83 |
| SRR519747 | 6 dpf | VD3 treatment | 156.88 |
| SRR1621206 | embryo | normal | 105.07 |
| SRR519737 | 2 dpf | VD3 treatment | 88.26 |
| SRR519717 | 2 dpf | FETOH treatment | 80.70 |
| SRR519742 | 4 dpf | VD3 treatment | 79.23 |
| SRR372793 | shield | normal | 78.44 |
Express in tissues
Correlated coding gene
Download
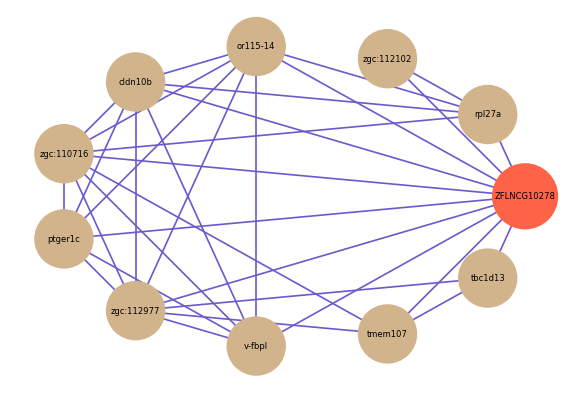
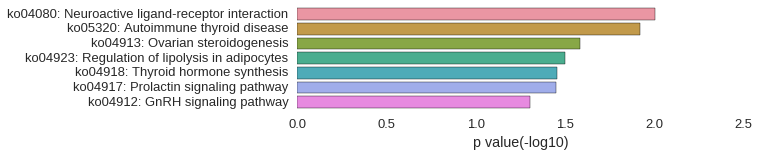
| gene | correlation coefficent |
|---|---|
| rpl27a | 0.58 |
| zgc:112102 | 0.55 |
| ptger1c | 0.55 |
| or115-14 | 0.55 |
| zgc:112977 | 0.54 |
| tmem107 | 0.54 |
| cldn10b | 0.53 |
| tbc1d13 | 0.53 |
| v-fbpl | 0.52 |
| zgc:110716 | 0.52 |
Gene Ontology
Download
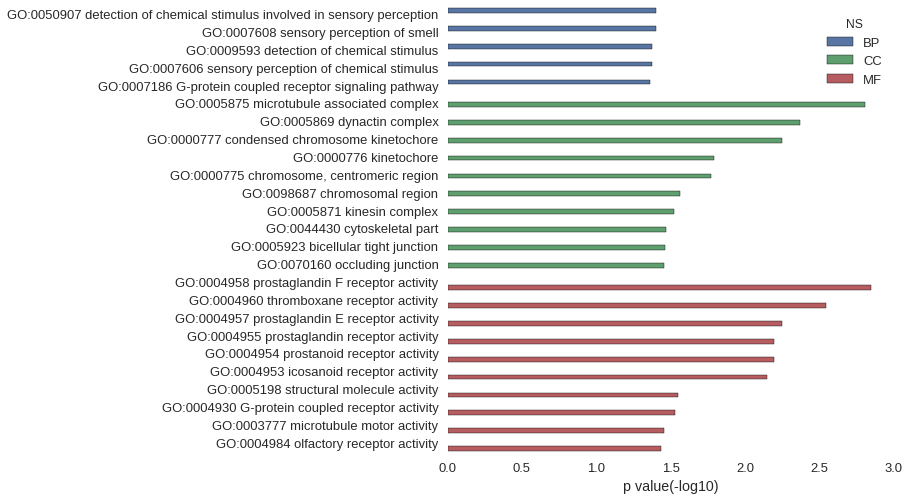
| GO | P value |
|---|---|
| GO:0050907 | 3.98e-02 |
| GO:0007608 | 3.98e-02 |
| GO:0009593 | 4.25e-02 |
| GO:0007606 | 4.25e-02 |
| GO:0007186 | 4.36e-02 |
| GO:0005875 | 1.57e-03 |
| GO:0005869 | 4.26e-03 |
| GO:0000777 | 5.67e-03 |
| GO:0000776 | 1.62e-02 |
| GO:0000775 | 1.69e-02 |
| GO:0098687 | 2.74e-02 |
| GO:0005871 | 3.01e-02 |
| GO:0044430 | 3.38e-02 |
| GO:0005923 | 3.43e-02 |
| GO:0070160 | 3.50e-02 |
| GO:0004958 | 1.42e-03 |
| GO:0004960 | 2.84e-03 |
| GO:0004957 | 5.67e-03 |
| GO:0004955 | 6.38e-03 |
| GO:0004954 | 6.38e-03 |
| GO:0004953 | 7.09e-03 |
| GO:0005198 | 2.84e-02 |
| GO:0004930 | 2.96e-02 |
| GO:0003777 | 3.50e-02 |
| GO:0004984 | 3.70e-02 |
KEGG Pathway
Download
Conservation
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10278
CGAGATATCCGCGTCATCATTCAGGCGGGGTTTATAAGAAGATGGGCGGGGTCTGCGCTGAGAGGGCGTG
TCTTCATTGTGTGTCGTGTCATGTCTGCTGTCTTTCTTCTGCTGCTGTTTTCTGTCGTGCCACGTTGATC
TAGCGATCGTCAAACTGCAGGTGAGAACGCAGATGACGGACTGCTGCTTCTGTCCTCACACACACACTCT
CTCTCAGTGAAGTGTGTTCATGGCTAGACTCTCGCTTTCCCGCTTTCCGAACTTCAGGTGTTGTTGTGTG
TTAGCAATGCTAGCTGCTCTGAGGTGCTCGACATTCAGATCTGCGCCCAGTCAGGT
CGAGATATCCGCGTCATCATTCAGGCGGGGTTTATAAGAAGATGGGCGGGGTCTGCGCTGAGAGGGCGTG
TCTTCATTGTGTGTCGTGTCATGTCTGCTGTCTTTCTTCTGCTGCTGTTTTCTGTCGTGCCACGTTGATC
TAGCGATCGTCAAACTGCAGGTGAGAACGCAGATGACGGACTGCTGCTTCTGTCCTCACACACACACTCT
CTCTCAGTGAAGTGTGTTCATGGCTAGACTCTCGCTTTCCCGCTTTCCGAACTTCAGGTGTTGTTGTGTG
TTAGCAATGCTAGCTGCTCTGAGGTGCTCGACATTCAGATCTGCGCCCAGTCAGGT