ZFLNCG10356
Basic Information
Genome Browser
Express profile
Download
Express in top 10 samples
| Sample | Tissue | Condition | FPKM |
|---|---|---|---|
| SRR1004789 | larvae | impdh2 morpholino | 371.23 |
| SRR1004787 | larvae | impdh1a morpholino | 350.40 |
| SRR1005530 | embryo | RPL11 morpholino | 300.26 |
| SRR1534349 | larvae | DMSO vehicle | 192.09 |
| SRR1004788 | larvae | impdh1b morpholino | 159.85 |
| SRR535943 | larvae | vangl mut | 137.41 |
| SRR1004785 | larvae | normal | 131.88 |
| SRR038624 | embryo | traf6 morpholino and bacterial infection | 131.75 |
| SRR038625 | embryo | traf6 morpholino | 127.34 |
| SRR535926 | larvae | normal | 119.17 |
Express in tissues
Correlated coding gene
Download
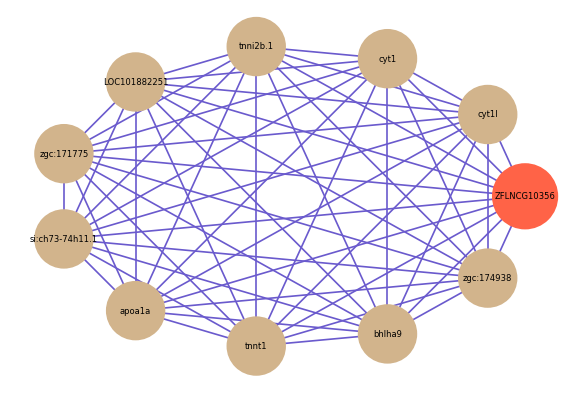
| gene | correlation coefficent |
|---|---|
| cyt1l | 0.76 |
| cyt1 | 0.75 |
| tnni2b.1 | 0.67 |
| LOC101882251 | 0.66 |
| zgc:171775 | 0.65 |
| si:ch73-74h11.1 | 0.64 |
| apoa1a | 0.64 |
| tnnt1 | 0.64 |
| bhlha9 | 0.64 |
| zgc:174938 | 0.63 |
Gene Ontology
Download
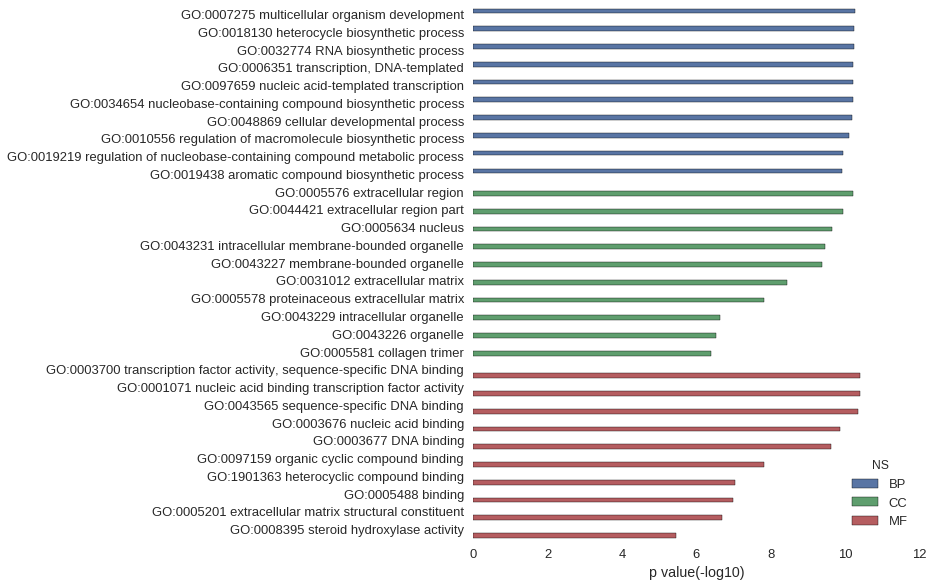
| GO | P value |
|---|---|
| GO:0007275 | 5.39e-11 |
| GO:0018130 | 5.84e-11 |
| GO:0032774 | 5.93e-11 |
| GO:0006351 | 6.08e-11 |
| GO:0097659 | 6.08e-11 |
| GO:0034654 | 6.19e-11 |
| GO:0048869 | 6.61e-11 |
| GO:0010556 | 7.85e-11 |
| GO:0019219 | 1.11e-10 |
| GO:0019438 | 1.19e-10 |
| GO:0005576 | 6.00e-11 |
| GO:0044421 | 1.17e-10 |
| GO:0005634 | 2.31e-10 |
| GO:0043231 | 3.39e-10 |
| GO:0043227 | 4.12e-10 |
| GO:0031012 | 3.74e-09 |
| GO:0005578 | 1.56e-08 |
| GO:0043229 | 2.38e-07 |
| GO:0043226 | 2.94e-07 |
| GO:0005581 | 4.13e-07 |
| GO:0003700 | 4.07e-11 |
| GO:0001071 | 4.07e-11 |
| GO:0043565 | 4.45e-11 |
| GO:0003676 | 1.34e-10 |
| GO:0003677 | 2.35e-10 |
| GO:0097159 | 1.50e-08 |
| GO:1901363 | 8.99e-08 |
| GO:0005488 | 1.03e-07 |
| GO:0005201 | 1.99e-07 |
| GO:0008395 | 3.49e-06 |
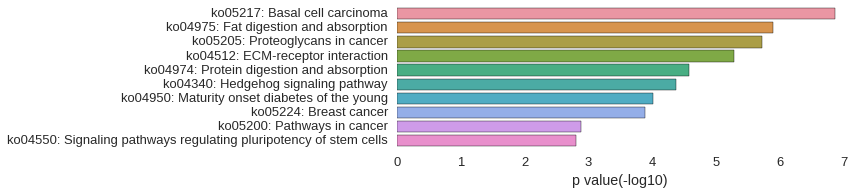
KEGG Pathway
Download
Conservation
| Zebrafish lncRNA Transcript | Human lncRNA Transcript | Mouse lncRNA Transcript | Methods |
|---|---|---|---|
| ZFLNCT16042 | ENST00000411759; NONHSAT176187; NONHSAT176254; NONHSAT176198; NONHSAT176955; NONHSAT176191; NONHSAT053996; NONHSAT176199; NONHSAT176193; NONHSAT176189; NONHSAT176188; NONHSAT053631; NONHSAT163560; NONHSAT176196; NONHSAT053995; NONHSAT176190; NONHSAT175207; NONHSAT176195; NONHSAT175206; NONHSAT053988; NONHSAT053990; NONHSAT176194; NONHSAT176255; NONHSAT176192; | Collinearity with conserved coding gene |
OMIM
OMIM of this lncRNA gene hava does not exist!
Sequence Download
>ZFLNCG10356
AGGAAGATCAGTTCTTCTTTCAGACCCTCGATCTGCAACTCCAGGTCAGATCTGGTCATTGTGAGCTCAT
CCAGCACCTTCCTCAAGCCAACAATGTCTGCTTCCACAGACTGCCTCATGGTCAGCTCATTCTCATACCT
GCACAGAAATAAAATCAATGTAAATGTTTTGGATCTATTATAGATTTTTTAAGTGTTTTTTTAATTGTGC
AAGTCTTACAACAGTAGACTTACTTGACTCTGAAGTCATCTGCAGCCAGCTTGGCATTGTCAATGCTGAG
GTAGATGCCTCCATTTACACCAGTAGCATGCAGGATCTTTAAGGACAAATGCAATGATTTTCTTAAATAA
TATGAAGCATCAAACCTTCATAAAAAGATATTTGGAATTTGTTCATTTGGAAATGCAATGATTAAATTAA
TCACACATTCAAAAGTTGTCACATACCTTTGCCTGGAGGTCACTGATTGTTGCATAGTAGGCACTGTAGT
CACGAGCATTAGGCGTGGCCTTGCTGTCCAGGAACTGGCGGATCTTCAGCTCAAGATCTGCATTGGCCTT
CTCCAGGCTGCGCACCTTCTCCAGGTAGGAGGCCAGACGGTCATTGAGGTTTTGCATGGTGGCTTTCTCA
TTTGTTGACACGTCAATGGCATCAGACAGGTTGAAGCCAGCACCAGCACCACCACCAAATCCTGCGCCGC
CACCAGCAGAGAAGGATCTGGAAGCAGAAGAACTGGAGATGCGAACACCGGATCCTCCTGCTCCTCCATA
GACACTACCAGCACGCACTCCTGAGACACGACCGGCTCCACCACCGACAGAGAGGCGGGAAGATCCTCCT
GAGGACATGCGGATGGATGAAGAAGACATCATTTTGAAGGAGTCTTGCTGTCGTCGTCTGCTCTACCTGG
AGAAGGAAGTGAGCGGATGAGTGAGGAGAAGACG
AGGAAGATCAGTTCTTCTTTCAGACCCTCGATCTGCAACTCCAGGTCAGATCTGGTCATTGTGAGCTCAT
CCAGCACCTTCCTCAAGCCAACAATGTCTGCTTCCACAGACTGCCTCATGGTCAGCTCATTCTCATACCT
GCACAGAAATAAAATCAATGTAAATGTTTTGGATCTATTATAGATTTTTTAAGTGTTTTTTTAATTGTGC
AAGTCTTACAACAGTAGACTTACTTGACTCTGAAGTCATCTGCAGCCAGCTTGGCATTGTCAATGCTGAG
GTAGATGCCTCCATTTACACCAGTAGCATGCAGGATCTTTAAGGACAAATGCAATGATTTTCTTAAATAA
TATGAAGCATCAAACCTTCATAAAAAGATATTTGGAATTTGTTCATTTGGAAATGCAATGATTAAATTAA
TCACACATTCAAAAGTTGTCACATACCTTTGCCTGGAGGTCACTGATTGTTGCATAGTAGGCACTGTAGT
CACGAGCATTAGGCGTGGCCTTGCTGTCCAGGAACTGGCGGATCTTCAGCTCAAGATCTGCATTGGCCTT
CTCCAGGCTGCGCACCTTCTCCAGGTAGGAGGCCAGACGGTCATTGAGGTTTTGCATGGTGGCTTTCTCA
TTTGTTGACACGTCAATGGCATCAGACAGGTTGAAGCCAGCACCAGCACCACCACCAAATCCTGCGCCGC
CACCAGCAGAGAAGGATCTGGAAGCAGAAGAACTGGAGATGCGAACACCGGATCCTCCTGCTCCTCCATA
GACACTACCAGCACGCACTCCTGAGACACGACCGGCTCCACCACCGACAGAGAGGCGGGAAGATCCTCCT
GAGGACATGCGGATGGATGAAGAAGACATCATTTTGAAGGAGTCTTGCTGTCGTCGTCTGCTCTACCTGG
AGAAGGAAGTGAGCGGATGAGTGAGGAGAAGACG